Martial Marbouty
@mmarbout.bsky.social
Organizations, structures and dynamics of microbial communities. Phages lover. CNRS researcher. Developer of HiC technics and bioinformatic pipelines applied to metagenomes. Opinions are my own.
Thanks Meren !! Coming from you … I am honoured !!
September 19, 2025 at 5:30 PM
Thanks Meren !! Coming from you … I am honoured !!
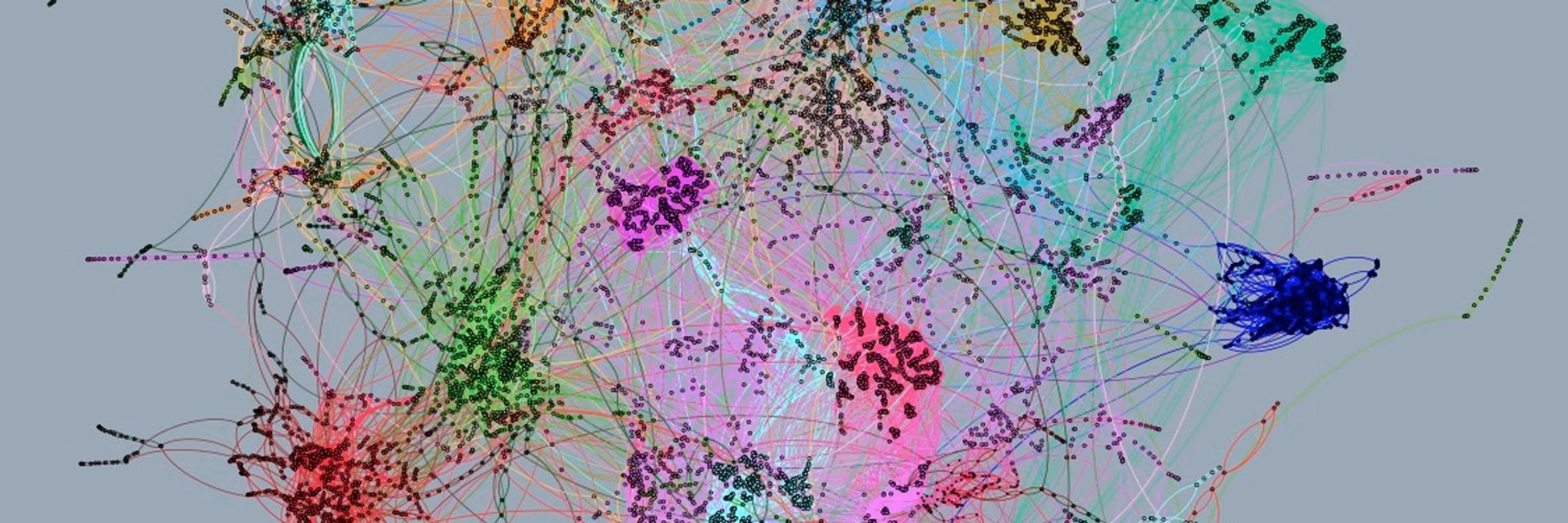
Our work show that phages with a broad host ranges are common across ecosystems. It also highlights the interest of using HiC to study single metagenomic samples.
Congrats to all authors !! 🎉
Congrats to all authors !! 🎉
September 19, 2025 at 9:47 AM
Our work show that phages with a broad host ranges are common across ecosystems. It also highlights the interest of using HiC to study single metagenomic samples.
Congrats to all authors !! 🎉
Congrats to all authors !! 🎉
We then show that among the phages with an assigned host, a substantial proportion of them interact with several, and sometimes distantly related, hosts.
September 19, 2025 at 9:45 AM
We then show that among the phages with an assigned host, a substantial proportion of them interact with several, and sometimes distantly related, hosts.
We have been able to reconstruct numerous good quality viral genomes up to 1Mb from diverse families such as Megaviricetes or Caudoviricetes.
September 19, 2025 at 9:43 AM
We have been able to reconstruct numerous good quality viral genomes up to 1Mb from diverse families such as Megaviricetes or Caudoviricetes.
We then apply our approach on public and newly generated metaHiC datasets (111 in total) from different ecosystems to explore the virosphere of these communities.
September 19, 2025 at 9:41 AM
We then apply our approach on public and newly generated metaHiC datasets (111 in total) from different ecosystems to explore the virosphere of these communities.
We first used a synthetic community composed of various microorganisms and mobile genetic elements to develop a new version of our MetaTOR pipeline allowing to reconstruct virus genomes and identify their hosts in microbial communities.
github.com/koszullab/me...
github.com/koszullab/me...

GitHub - koszullab/metaTOR: Metagenomic binning based on Hi-C data
Metagenomic binning based on Hi-C data. Contribute to koszullab/metaTOR development by creating an account on GitHub.
github.com
September 19, 2025 at 9:38 AM
We first used a synthetic community composed of various microorganisms and mobile genetic elements to develop a new version of our MetaTOR pipeline allowing to reconstruct virus genomes and identify their hosts in microbial communities.
github.com/koszullab/me...
github.com/koszullab/me...
Reposted by Martial Marbouty
Reportage de @celineloozen.bsky.social > Itw de Martial Marbouty, chercheur CNRS au laboratoire Régulation Spatiale des Génomes de l'Institut Pasteur sur l’étude des interactions phages-bactéries tinyurl.com/m9uxwjct #ScienceCQFD

Comment détourner les bactériophages à des fins thérapeutiques.
Dans nos organismes, les relations entre bactéries et phages tissent un réseau complexe dont l'analyse ouvre des thérapies innovantes, notamment pour réguler le microbiote intestinal.
tinyurl.com
May 21, 2025 at 2:34 PM
Reportage de @celineloozen.bsky.social > Itw de Martial Marbouty, chercheur CNRS au laboratoire Régulation Spatiale des Génomes de l'Institut Pasteur sur l’étude des interactions phages-bactéries tinyurl.com/m9uxwjct #ScienceCQFD
Great science, great discussions, amazing people… a real pleasure to be here !!
February 27, 2025 at 12:04 PM
Great science, great discussions, amazing people… a real pleasure to be here !!

