

www.science.org/doi/10.1126/...
Preprint: www.biorxiv.org/content/10.1...
Preprint: www.biorxiv.org/content/10.1...
Read more: bit.ly/3SCZQiM
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.nature.com/articles/s41...
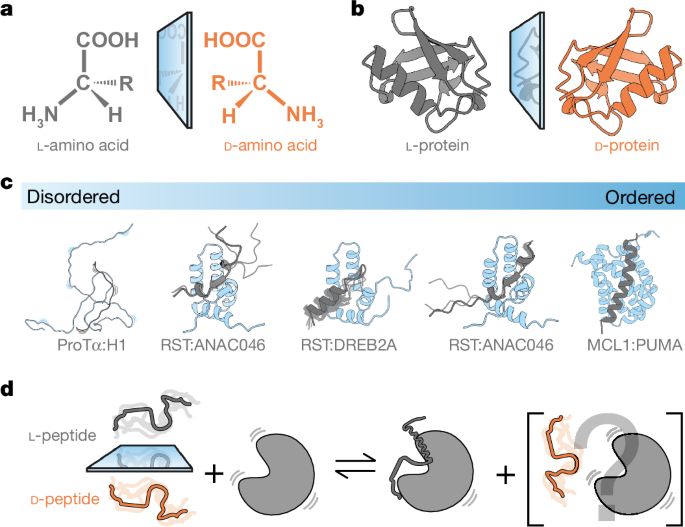
www.nature.com/articles/s41...
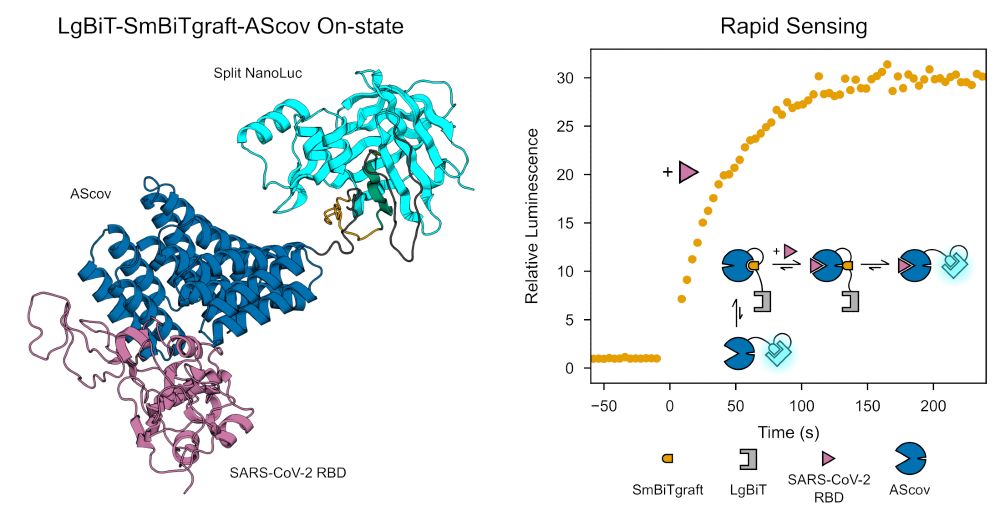
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

