
Preprint: www.biorxiv.org/content/10.1...
The first preprint out of my lab! We joined forces with @kinasekid.bsky.social @jasonzxzhang.bsky.social and David Baker to study protein phosphorylation! Congrats to Isabella from my lab on her first first author paper! tinyurl.com/43jwwfua
The first preprint out of my lab! We joined forces with @kinasekid.bsky.social @jasonzxzhang.bsky.social and David Baker to study protein phosphorylation! Congrats to Isabella from my lab on her first first author paper! tinyurl.com/43jwwfua
The first preprint out of my lab! We joined forces with @kinasekid.bsky.social @jasonzxzhang.bsky.social and David Baker to study protein phosphorylation! Congrats to Isabella from my lab on her first first author paper! tinyurl.com/43jwwfua
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
🧬Code and notebooks will be released by the end of this week.
🎧Golden- Kpop Demon Hunters
Preprint: www.biorxiv.org/content/10.1...
Preprint: www.biorxiv.org/content/10.1...
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
www.youtube.com/live/z8NO4Bg...

www.youtube.com/live/z8NO4Bg...
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
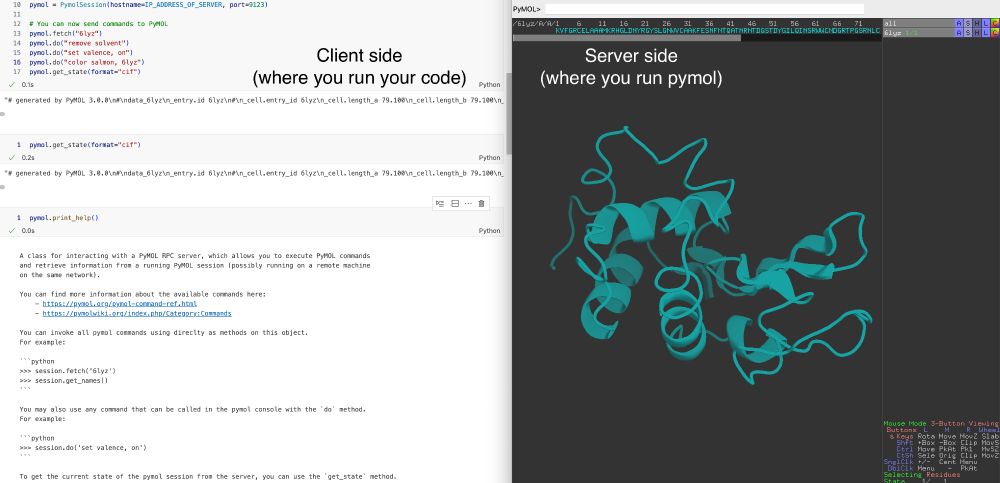
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
I wonder, is there any computational approach, that would find protein in PDB according to any arbitrary shape?
I wonder, is there any computational approach, that would find protein in PDB according to any arbitrary shape?
www.dropbox.com/scl/fi/6iny2...
www.dropbox.com/scl/fi/6iny2...
ProteinMPNN just doesn't like Chroma's backbones (poor prediction of proteinMPNN generated sequences by ESMFold). Interestingly, Chroma's own sequence design method (which was trained in the context of partially noise backbones) loves it! (1/3)
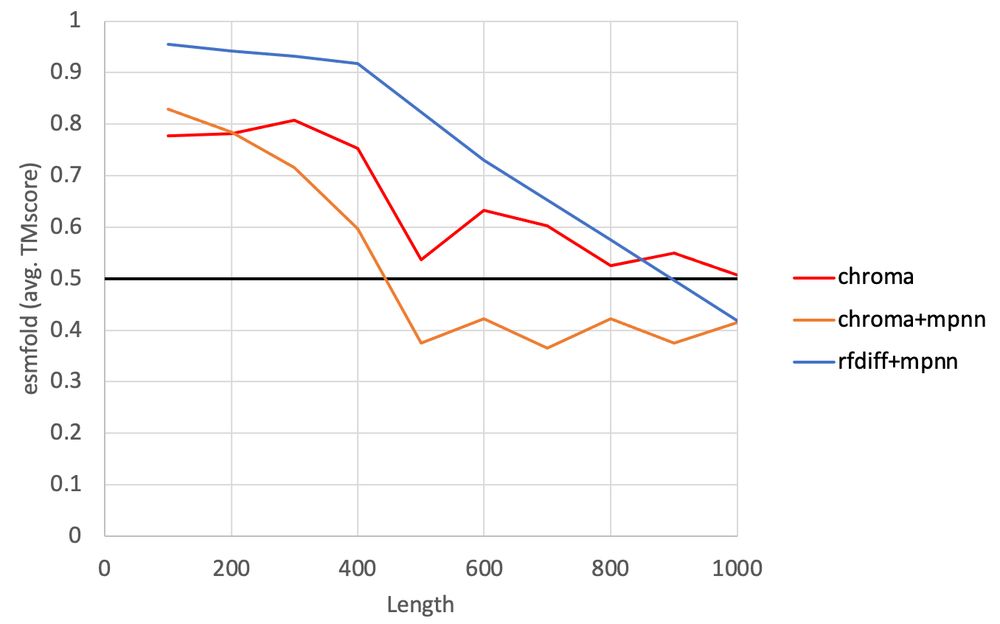
ProteinMPNN just doesn't like Chroma's backbones (poor prediction of proteinMPNN generated sequences by ESMFold). Interestingly, Chroma's own sequence design method (which was trained in the context of partially noise backbones) loves it! (1/3)
latest, in-development AlphaFold model" 🧪🧶🧬
DeepMind & Isomorphic Labs sharing some updates (but no code) on what is presumably alphafold 3, capable of modeling ligands, nucleic acids, antibody-antigen complexes etc
storage.googleapis.com/deepmind-med...

latest, in-development AlphaFold model" 🧪🧶🧬
DeepMind & Isomorphic Labs sharing some updates (but no code) on what is presumably alphafold 3, capable of modeling ligands, nucleic acids, antibody-antigen complexes etc
storage.googleapis.com/deepmind-med...

