
1/2
rachel.fast.ai/posts/2025-0...
1/2
rachel.fast.ai/posts/2025-0...
Predicts TF binding in target species via aligned sequence data distribution moments for cross-species generalization.
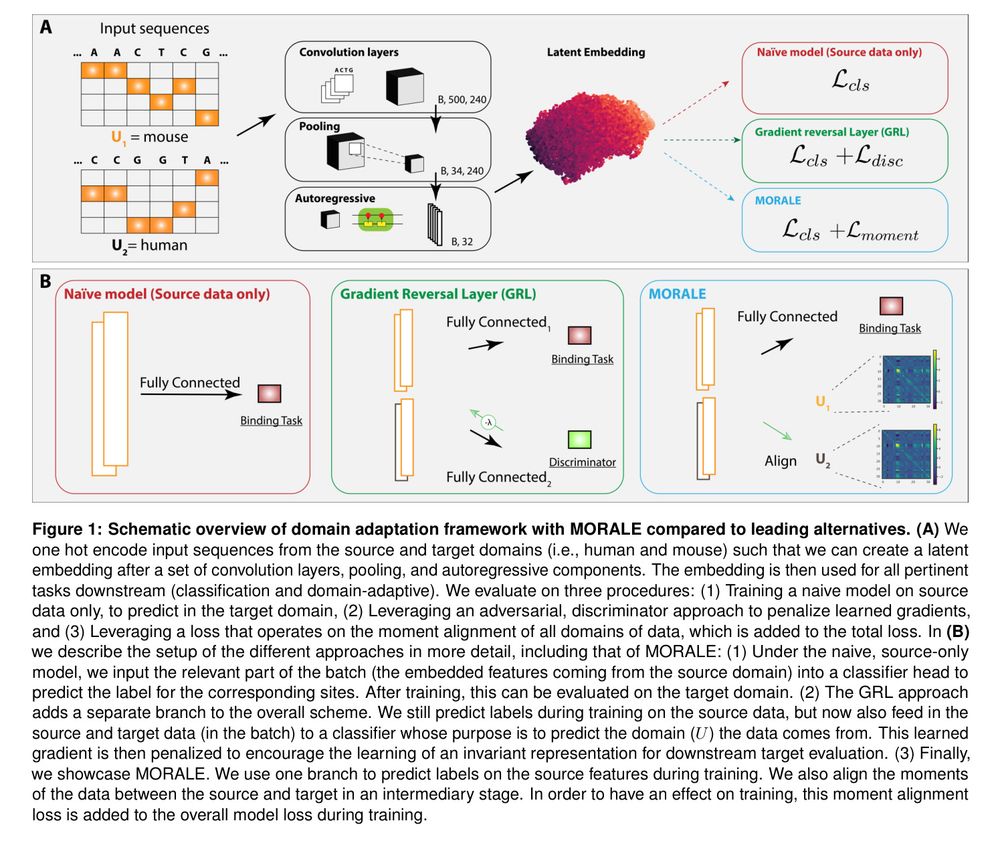
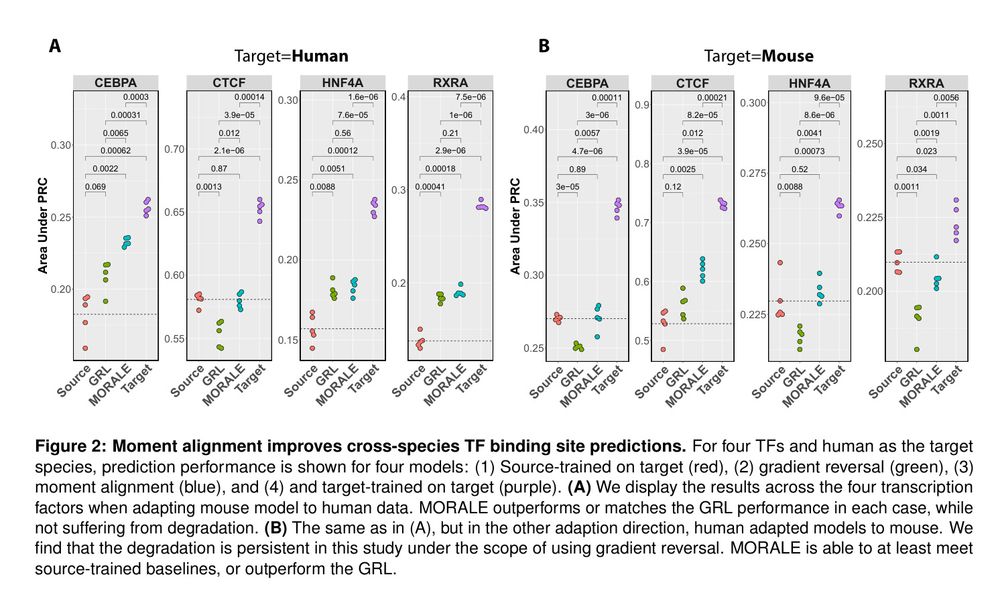
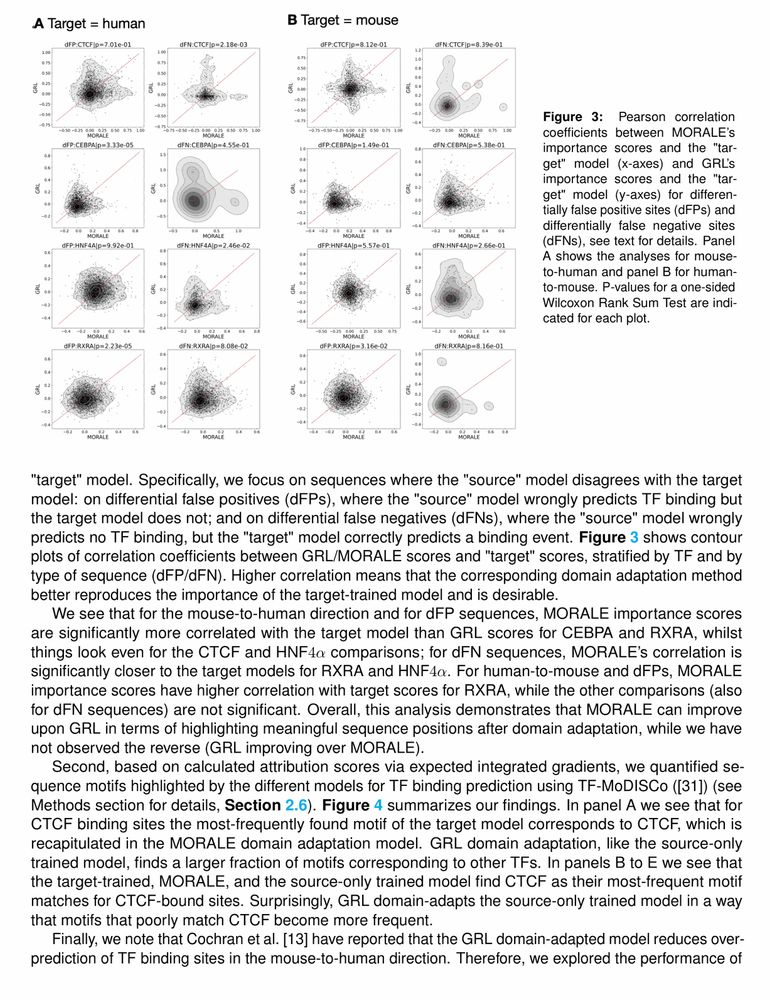
Predicts TF binding in target species via aligned sequence data distribution moments for cross-species generalization.
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...
Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.

Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.
Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

@lxsasse.bsky.social
@zmbh.uni-heidelberg.de
👉Advanced training strategies for genomic sequence-to-function models
📅 Wed March 5, 5:30pm CET
🧬 kipoi.org/seminar/
🦋 @kipoizoo.bsky.social
@lxsasse.bsky.social
@zmbh.uni-heidelberg.de
👉Advanced training strategies for genomic sequence-to-function models
📅 Wed March 5, 5:30pm CET
🧬 kipoi.org/seminar/
🦋 @kipoizoo.bsky.social
@charlesdanko.bsky.social
www.biorxiv.org/content/10.1...

@charlesdanko.bsky.social
www.biorxiv.org/content/10.1...
Deep reinforcement learning accurately partitions reads into haplotype sets. It uses fragment graphs and the max-cut problem for the reward objective.
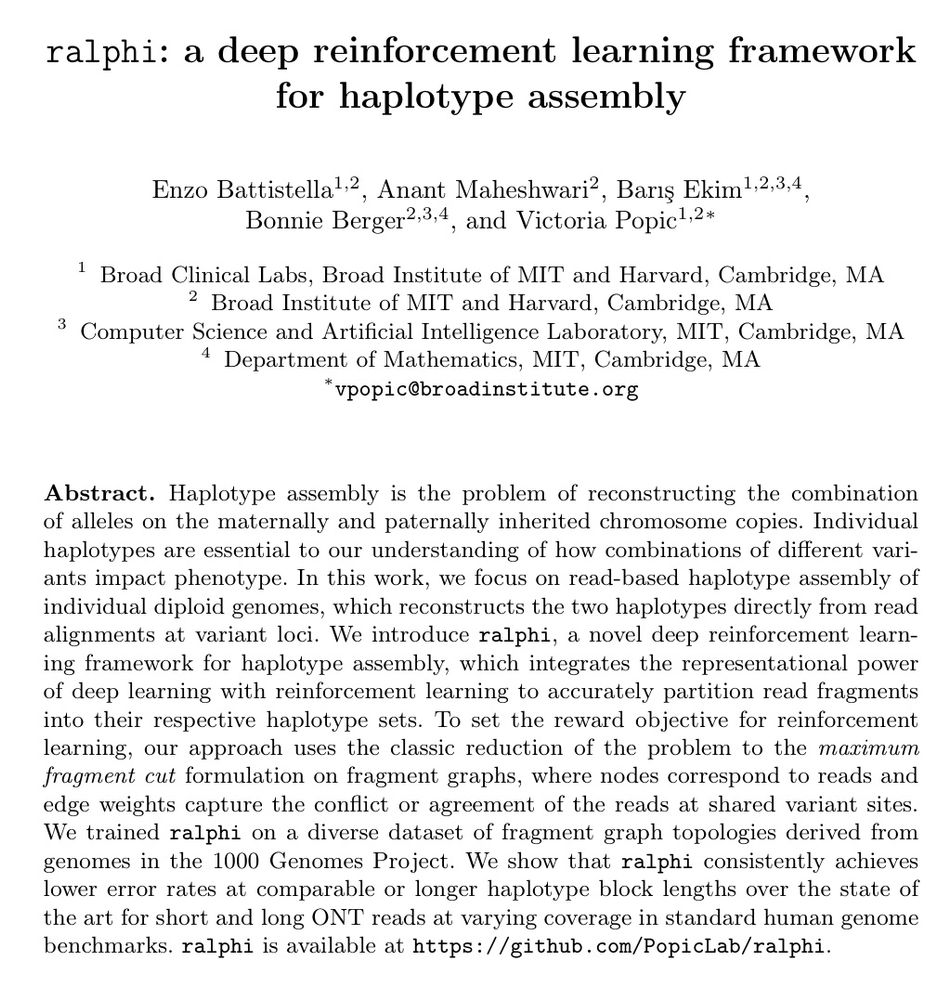
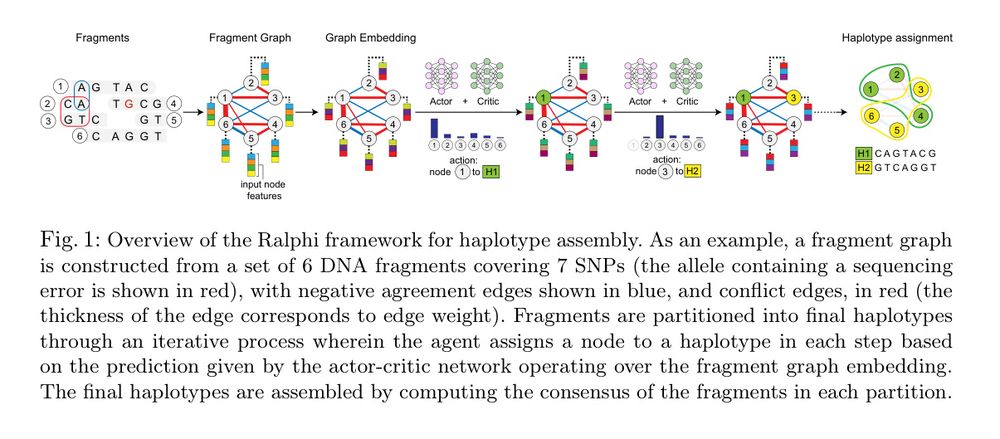
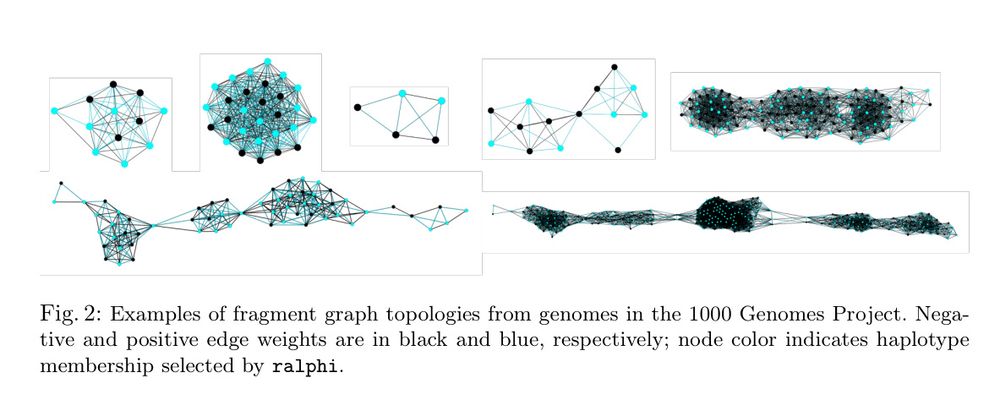
Deep reinforcement learning accurately partitions reads into haplotype sets. It uses fragment graphs and the max-cut problem for the reward objective.
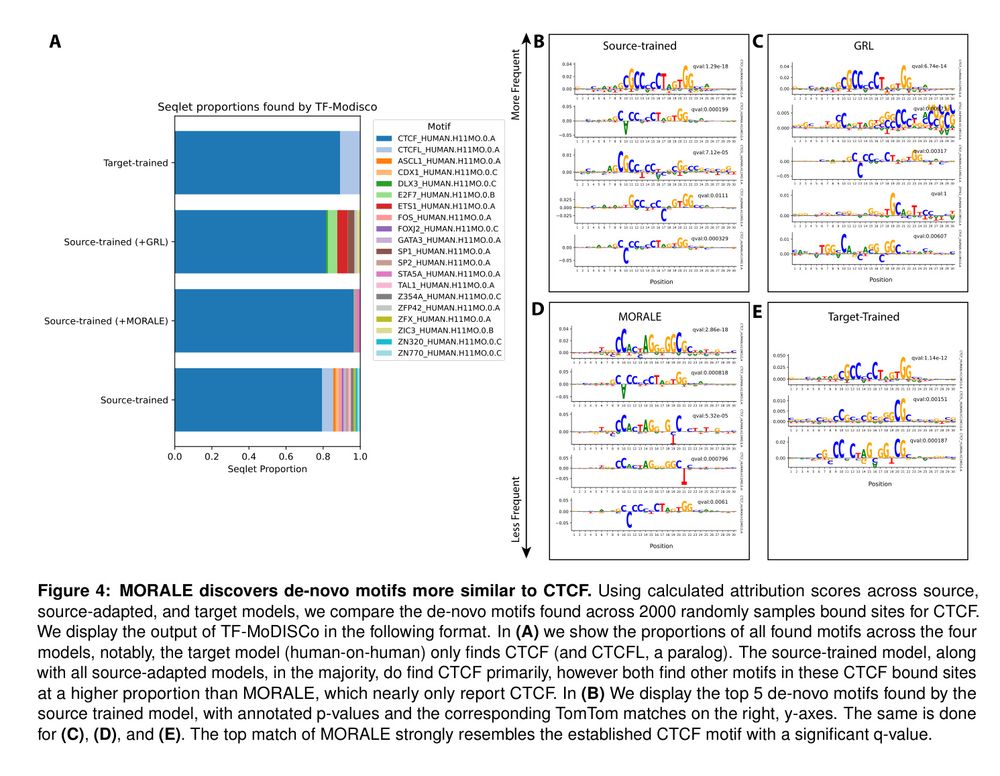
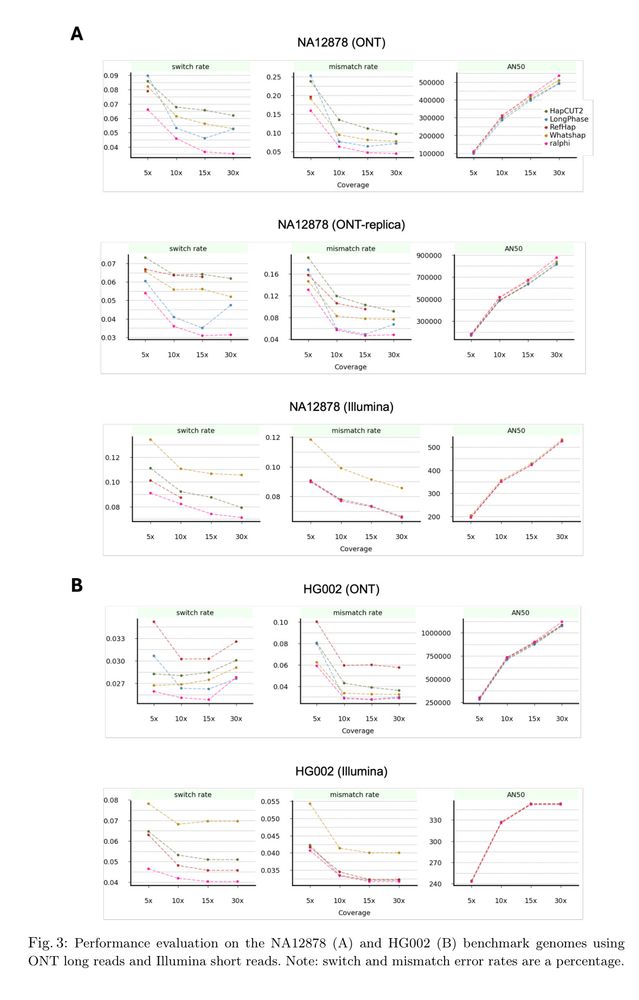
Models fail to gen. w/ individual var., personal genome training helps some individuals only.

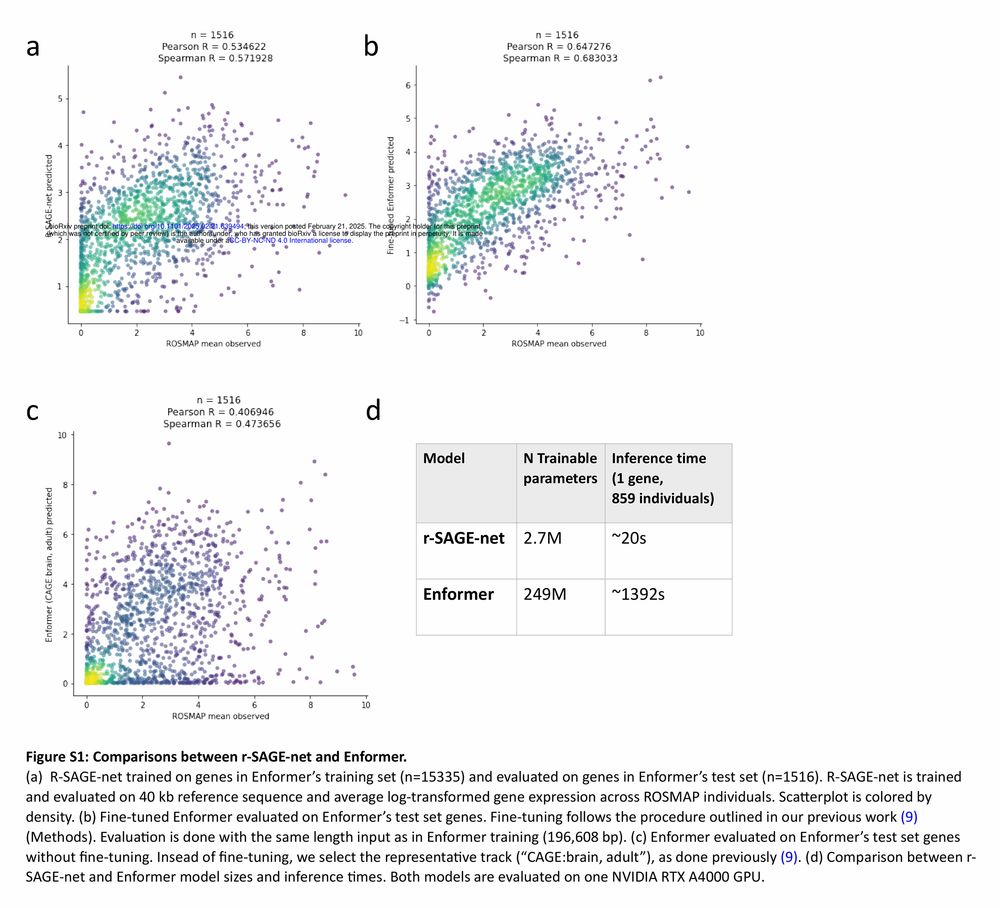
Models fail to gen. w/ individual var., personal genome training helps some individuals only.
Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...

Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...
Chromatin accessibility enhances sequence-to-expression models by focusing on open regions. Incorporating it improves predictions and reduces bias.




Chromatin accessibility enhances sequence-to-expression models by focusing on open regions. Incorporating it improves predictions and reduces bias.
TraitGym benchmarks reveal model-specific strengths in causal variant prediction for Mendelian/complex traits.




TraitGym benchmarks reveal model-specific strengths in causal variant prediction for Mendelian/complex traits.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
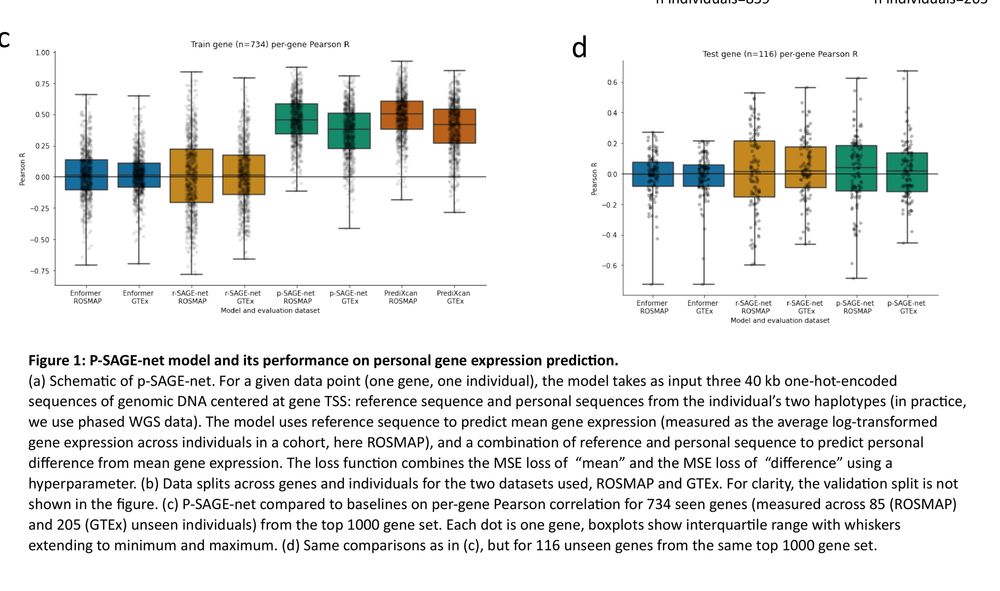
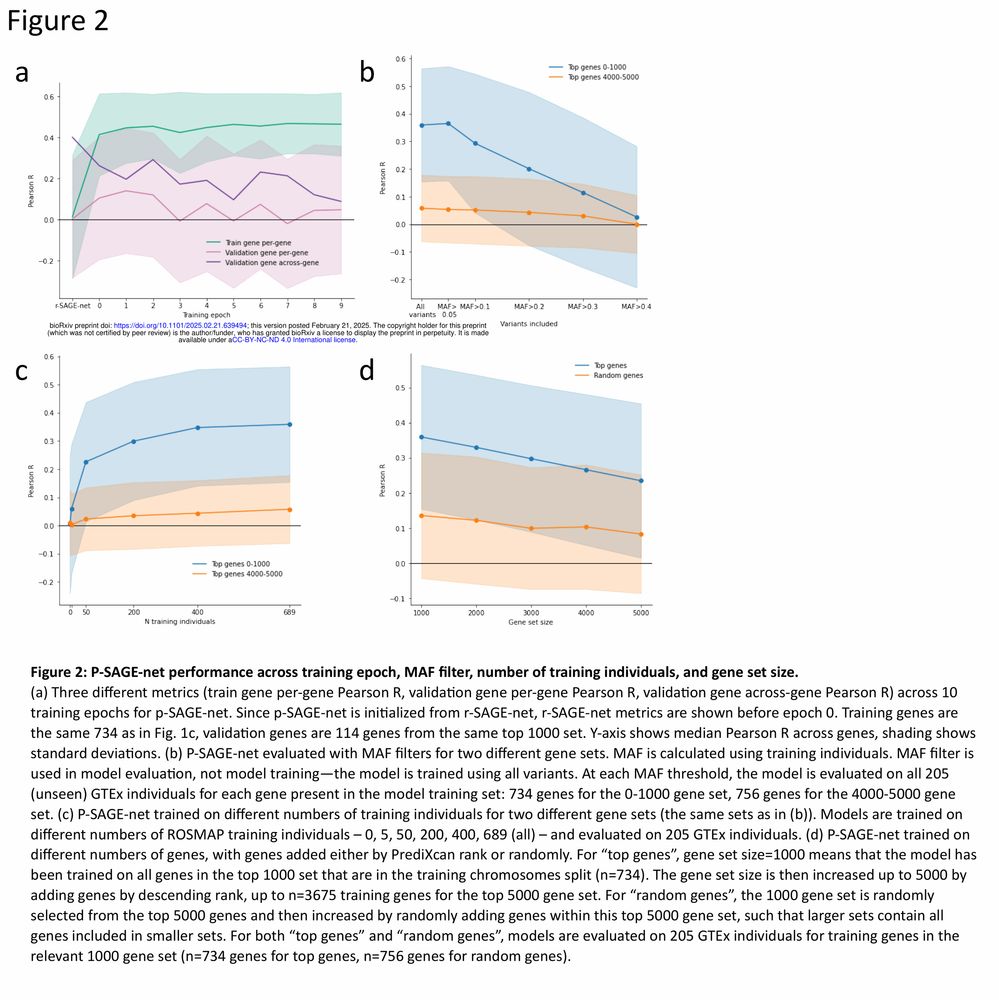
Might explain some of the discrepancies between QTL effect & personalized gene expression prediction performance by S2F models

Might explain some of the discrepancies between QTL effect & personalized gene expression prediction performance by S2F models




