Matthew J Shepherd
@matthewjshepherd.bsky.social
Postdoc at the University of Manchester, with Prof. Mike Brockhurst. Understanding and predicting the evolution of antimicrobial resistance in clinical Pseudomonas 🔬🦠
Pinned
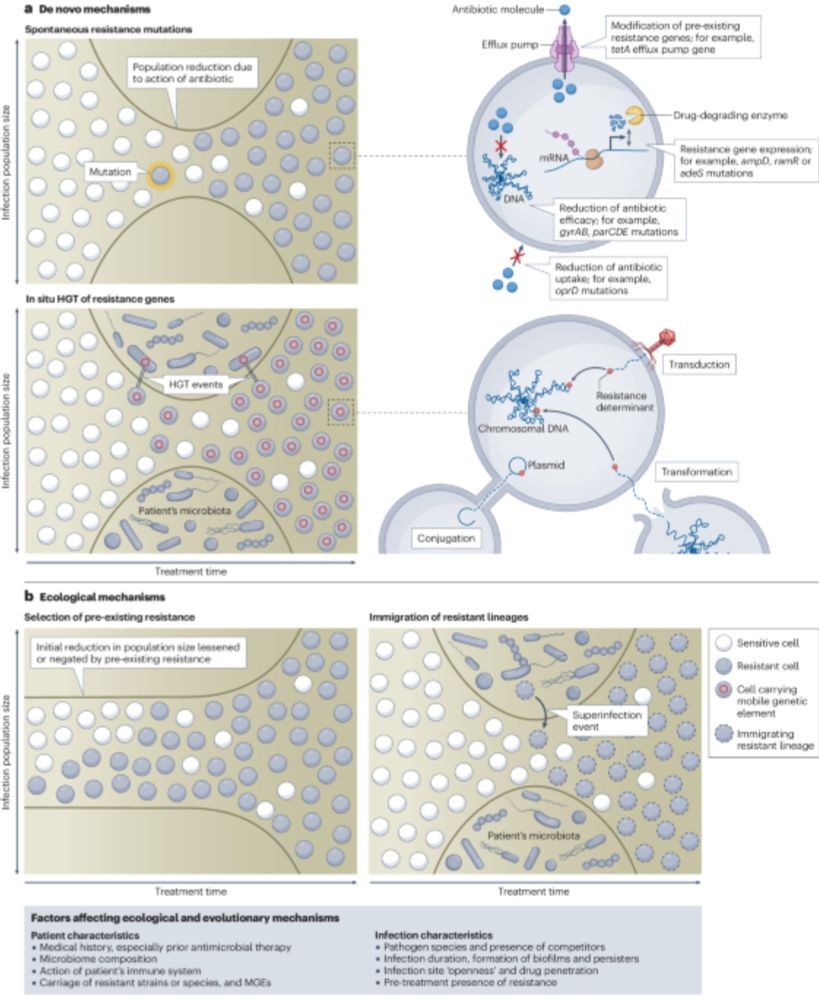
Ecological and evolutionary mechanisms driving within-patient emergence of antimicrobial resistance - Nature Reviews Microbiology
In this Review, Shepherd, Brockhurst and colleagues explore the clinical evidence in support of four major ecological and evolutionary mechanisms of within-patient antimicrobial resistance emergence i...
www.nature.com
Thrilled to share our new article now out in Nature Reviews Microbiology! We review how bacterial #AMR evolves within patients during #antibiotic therapy, highlighting the complexity of the problem and how to improve understanding in future work. 1/7 #MicroSky
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Matthew J Shepherd
We have a new preprint modeling the regulatory networks of efflux pumps to predict the evolution of multidrug resistance in P. aeruginosa. Also explore cross-resistance, collateral sensitivity & loss of resistance. With @suvamroy.bsky.social @elibbyscience.bsky.social www.biorxiv.org/content/10.1...

Forecasting multi-trait resistance evolution under antibiotic stress
Many bacteria rely on efflux pumps to survive antibiotic stress, and exposure to antibiotics often leads to mutations in pump genes or their regulators that increase pump expression. Predicting the sp...
www.biorxiv.org
October 20, 2025 at 6:21 AM
We have a new preprint modeling the regulatory networks of efflux pumps to predict the evolution of multidrug resistance in P. aeruginosa. Also explore cross-resistance, collateral sensitivity & loss of resistance. With @suvamroy.bsky.social @elibbyscience.bsky.social www.biorxiv.org/content/10.1...
Reposted by Matthew J Shepherd
Sharp global rise in antibiotic-resistant infections in hospitals, WHO finds

Sharp global rise in antibiotic-resistant infections in hospitals, WHO finds
Experts describe findings as deeply concerning and predict 70% increase in related deaths by 2050
Hospitals across the world have recorded an alarming rise in common infections that are resistant to antibiotics, with doctors saying the number of deaths driven by drug resistance will increase sharply in the years ahead.
One in six laboratory-confirmed bacterial infections were resistant to antibiotic treatments in 2023, with more than 40% of antibiotics losing potency against common blood, gut, urinary tract and sexually-transmitted infections between 2018 and 2023, records show. Continue reading...
www.theguardian.com
October 13, 2025 at 7:01 AM
Sharp global rise in antibiotic-resistant infections in hospitals, WHO finds
Reposted by Matthew J Shepherd
100k and rising!
This number will continue to grow.
Making hope normal again.
Join us.
join.greenparty.org.uk
This number will continue to grow.
Making hope normal again.
Join us.
join.greenparty.org.uk
🎉 We've done it. We've passed 100,000 Green Party members!
📈 But we're not stopping there.
💚 Let's make hope normal again! Join the Green Party today!
📈 But we're not stopping there.
💚 Let's make hope normal again! Join the Green Party today!
October 13, 2025 at 7:09 AM
100k and rising!
This number will continue to grow.
Making hope normal again.
Join us.
join.greenparty.org.uk
This number will continue to grow.
Making hope normal again.
Join us.
join.greenparty.org.uk
Reposted by Matthew J Shepherd
Reposted by Matthew J Shepherd
The evolutionary foundations of transcriptional regulation in animals www.nature.com/articles/s41... (read free: rdcu.be/evDcA) 🧬🖥️🧪
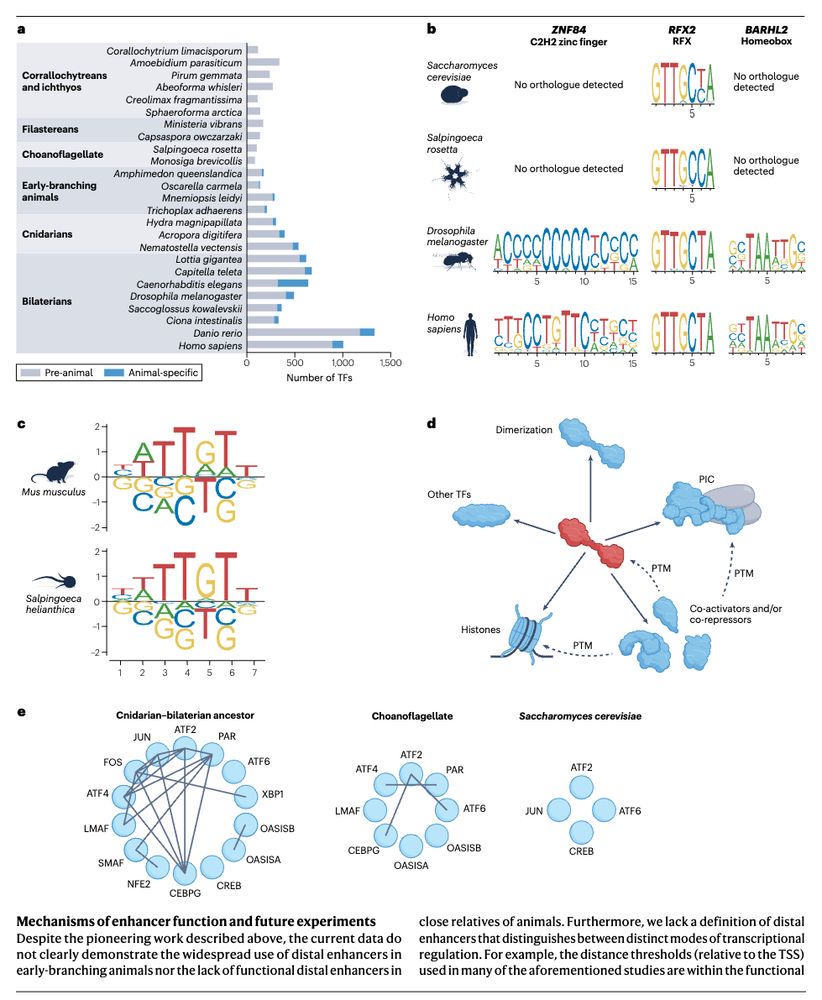
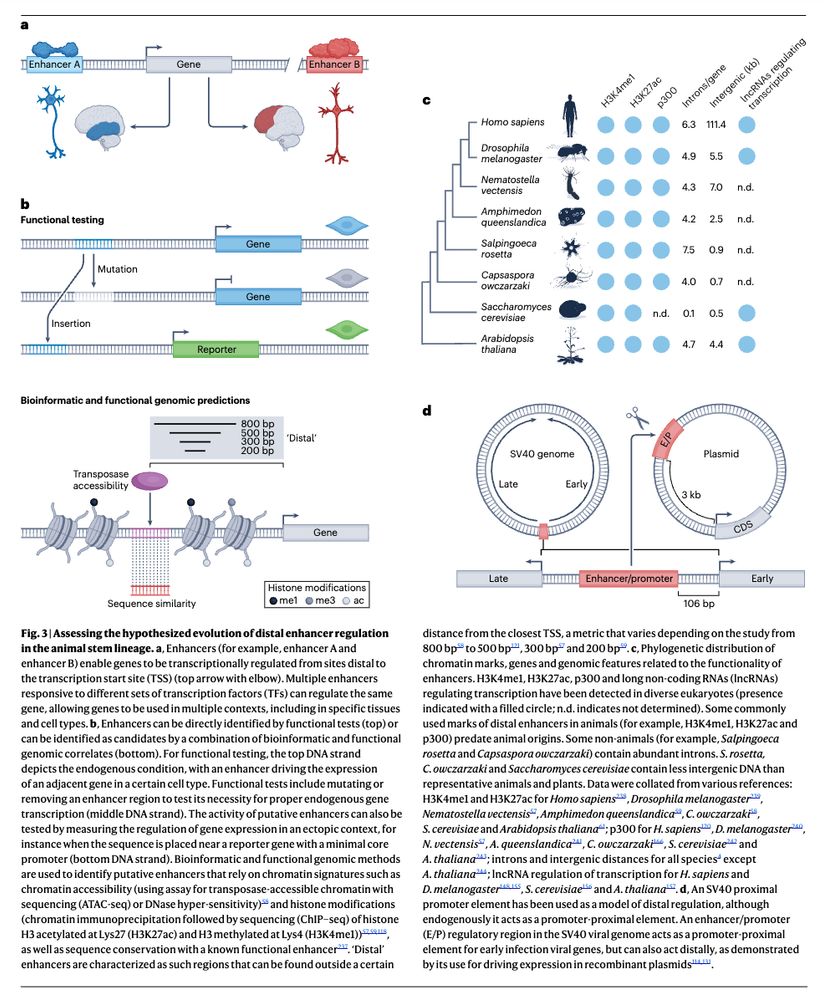
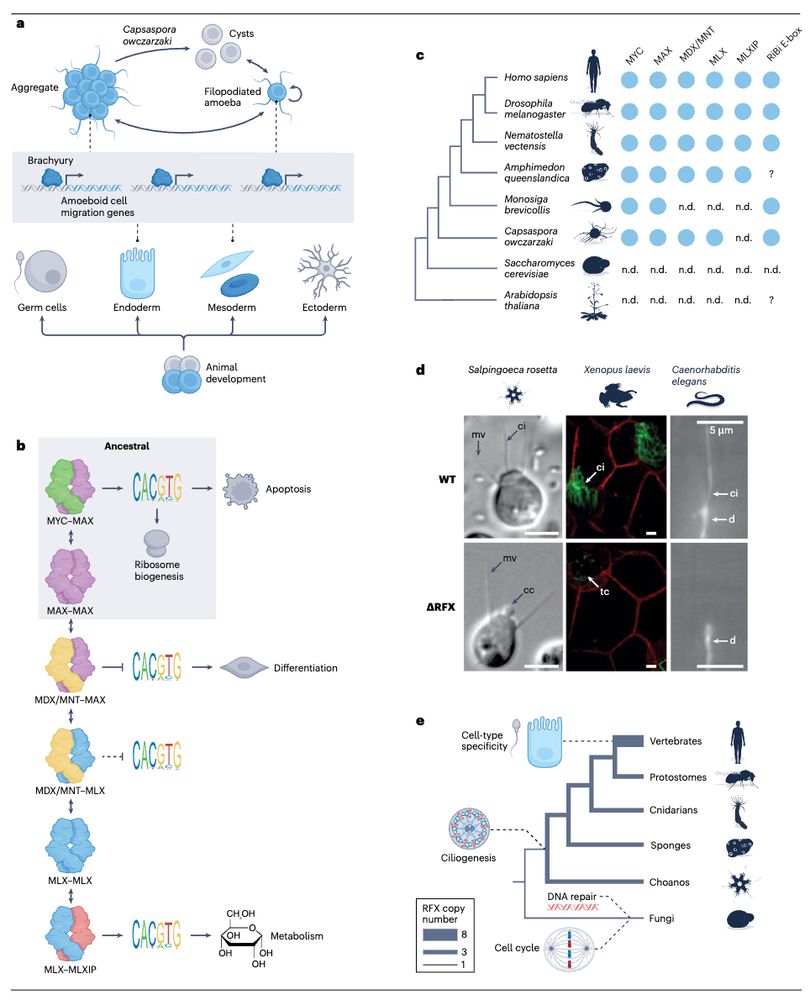
July 10, 2025 at 3:31 PM
The evolutionary foundations of transcriptional regulation in animals www.nature.com/articles/s41... (read free: rdcu.be/evDcA) 🧬🖥️🧪
Reposted by Matthew J Shepherd
The evolution of high-order genome architecture revealed from 1,000 species www.biorxiv.org/content/10.1... 🧬🖥️🧪 github.com/xjtu-omics/H...
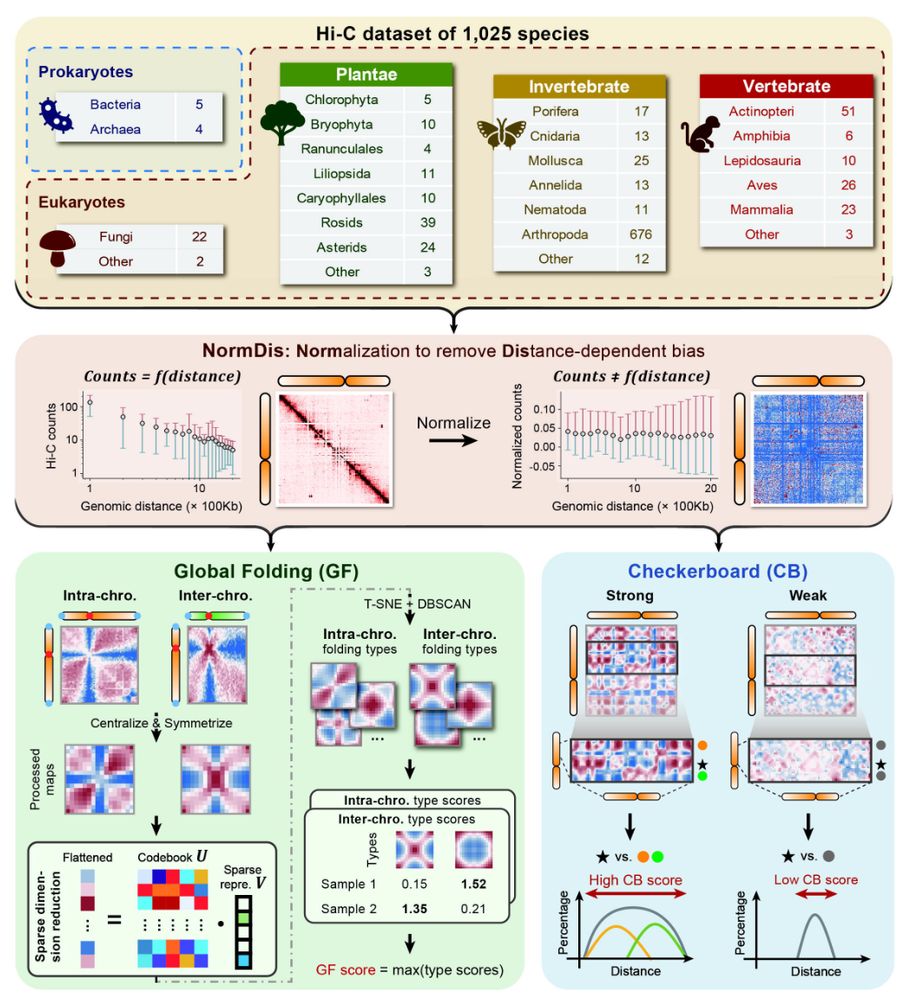
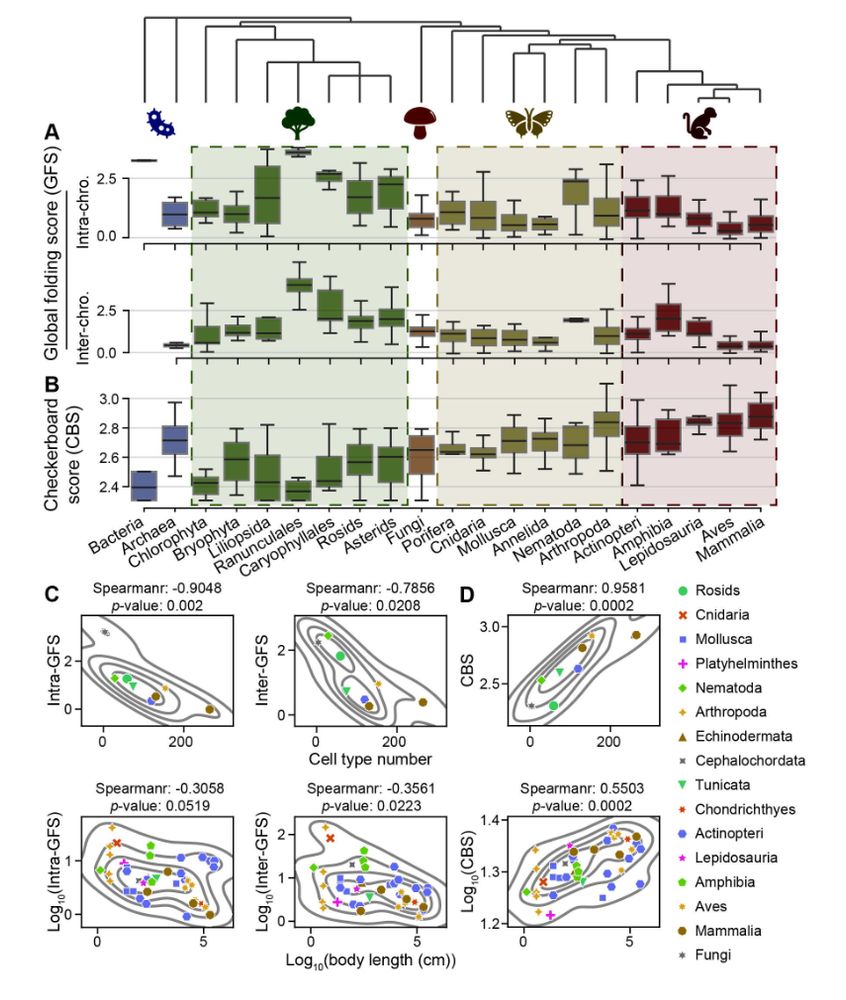
July 8, 2025 at 6:31 PM
The evolution of high-order genome architecture revealed from 1,000 species www.biorxiv.org/content/10.1... 🧬🖥️🧪 github.com/xjtu-omics/H...
Reposted by Matthew J Shepherd
Tissue destruction during food spoilage is associated with the formation of biofilms by Pseudomonas species https://www.biorxiv.org/content/10.1101/2025.07.11.664243v1
July 12, 2025 at 4:16 AM
Tissue destruction during food spoilage is associated with the formation of biofilms by Pseudomonas species https://www.biorxiv.org/content/10.1101/2025.07.11.664243v1
Reposted by Matthew J Shepherd
Patterns of compensatory mutations in rpoA/B/C genes of multidrug resistant M. tuberculosis in Uganda https://www.biorxiv.org/content/10.1101/2025.07.11.664293v1
July 12, 2025 at 5:17 AM
Patterns of compensatory mutations in rpoA/B/C genes of multidrug resistant M. tuberculosis in Uganda https://www.biorxiv.org/content/10.1101/2025.07.11.664293v1
Reposted by Matthew J Shepherd
1/27 We have a new paper out! Turns out that snowflake yeast have been hiding a secret from us - they've evolved a (very!) crude circulatory system. Not with blood vessels or a heart, but through spontaneous fluid flows powered by their metabolism. 🧪🔬
www.science.org/doi/full/10....
www.science.org/doi/full/10....
June 24, 2025 at 4:52 PM
1/27 We have a new paper out! Turns out that snowflake yeast have been hiding a secret from us - they've evolved a (very!) crude circulatory system. Not with blood vessels or a heart, but through spontaneous fluid flows powered by their metabolism. 🧪🔬
www.science.org/doi/full/10....
www.science.org/doi/full/10....
Reposted by Matthew J Shepherd
Higher-order epistasis drives evolutionary unpredictability toward novel antibiotic resistance https://www.biorxiv.org/content/10.1101/2025.07.08.663783v1
July 12, 2025 at 6:32 AM
Higher-order epistasis drives evolutionary unpredictability toward novel antibiotic resistance https://www.biorxiv.org/content/10.1101/2025.07.08.663783v1
Reposted by Matthew J Shepherd
A few hours left for you to sneak in a cheeky last minute abstract! ECRs, you know what to do…
#mevosky
#mevosky
One week left to submit your abstract to @microbiologysociety.org meeting on “Understanding and Predicting Microbial Evolutionary Dynamics” at Liverpool 26-27 November
Lots of spaces for ECR talks and posters!
microbiologysociety.org/event/societ...
Lots of spaces for ECR talks and posters!
microbiologysociety.org/event/societ...
microbiologysociety.org
July 7, 2025 at 4:25 PM
A few hours left for you to sneak in a cheeky last minute abstract! ECRs, you know what to do…
#mevosky
#mevosky
Reposted by Matthew J Shepherd
Fresh from peer review a new and improved version of @taoranfu.bsky.social’s paper out today in @asm.org mSystems
journals.asm.org/eprint/8NWNW...
journals.asm.org/eprint/8NWNW...
July 7, 2025 at 1:09 PM
Fresh from peer review a new and improved version of @taoranfu.bsky.social’s paper out today in @asm.org mSystems
journals.asm.org/eprint/8NWNW...
journals.asm.org/eprint/8NWNW...
Reposted by Matthew J Shepherd
Go Team! Deeply honoured that the MERMan lab reps won an @manchester.ac.uk "FMBH heroes" award - thanks @fbmh-uom.bsky.social and @mermanchester.bsky.social for nominating us ❤️❤️❤️


July 2, 2025 at 4:17 PM
Go Team! Deeply honoured that the MERMan lab reps won an @manchester.ac.uk "FMBH heroes" award - thanks @fbmh-uom.bsky.social and @mermanchester.bsky.social for nominating us ❤️❤️❤️
Reposted by Matthew J Shepherd
Curious about plasmid biology? Our latest paper is out now in Nature Communications! 🚨
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
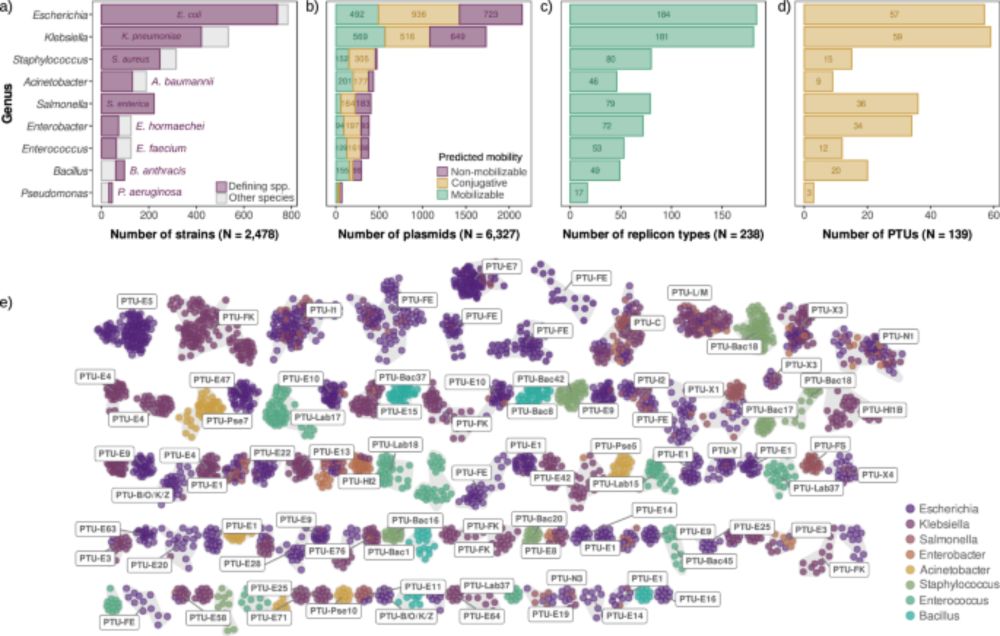
Universal rules govern plasmid copy number - Nature Communications
Plasmids exhibit a broad range of sizes and copies per cell, and these two parameters appear to be negatively correlated. Here, Ramiro-Martínez et al. analyse the copy number of thousands of diverse b...
doi.org
July 2, 2025 at 10:07 AM
Curious about plasmid biology? Our latest paper is out now in Nature Communications! 🚨
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
Reposted by Matthew J Shepherd
📣 Position available: Research tech in microbial experimental evolution! Working alongside postdoc Louise Flanagan (@flanagella.bsky.social) on understanding how Pseudomonas evolves resistance to antibiotics. 🦠🧫
Closing 9 July #Microsky
www.jobs.manchester.ac.uk/Job/JobDetai...
Closing 9 July #Microsky
www.jobs.manchester.ac.uk/Job/JobDetai...

Research Technician :Oxford Road
www.jobs.manchester.ac.uk
June 26, 2025 at 3:32 PM
📣 Position available: Research tech in microbial experimental evolution! Working alongside postdoc Louise Flanagan (@flanagella.bsky.social) on understanding how Pseudomonas evolves resistance to antibiotics. 🦠🧫
Closing 9 July #Microsky
www.jobs.manchester.ac.uk/Job/JobDetai...
Closing 9 July #Microsky
www.jobs.manchester.ac.uk/Job/JobDetai...
Reposted by Matthew J Shepherd
Mystique is here! When I started my position at @cmdi.bsky.social there were no readily available phage against my A. baumannii focal strain. The solution? Finding my own phage ofc. Mystique is a broad host range Acinetobacter phage, and I'm thrilled to see this work out today #PhageSky 🧪🦠
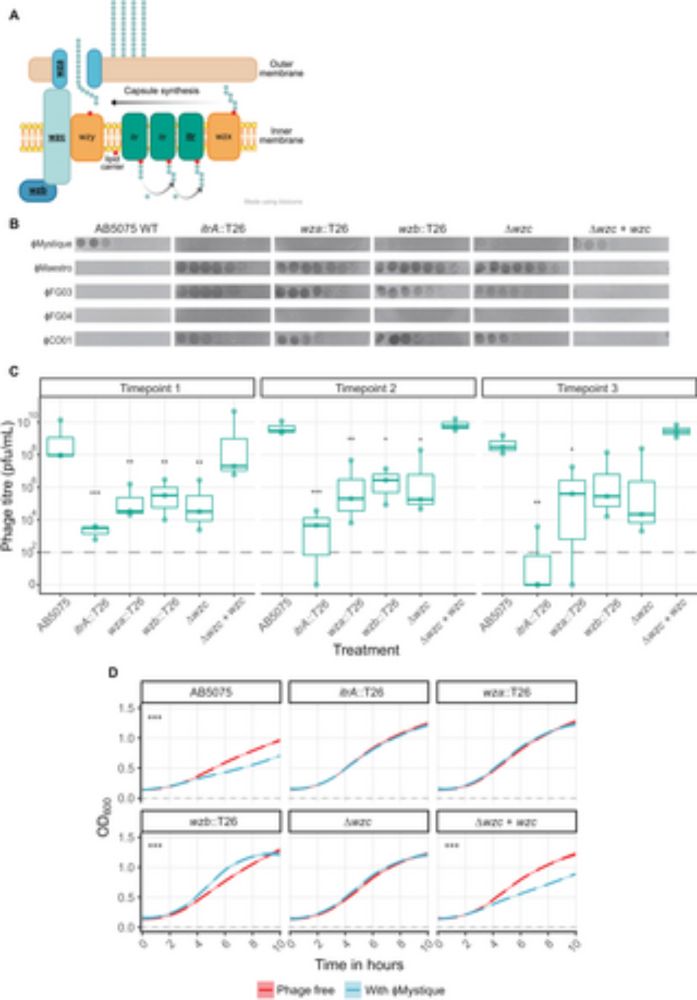
Mystique, a broad host range Acinetobacter phage, reveals the impact of culturing conditions on phage isolation and infectivity
Author summary Bacterial infections caused by Acinetobacter baumannii are a major global health concern due to high antibiotic resistance, earning it a critical priority pathogen ranking by the WHO. P...
journals.plos.org
April 10, 2025 at 7:59 PM
Mystique is here! When I started my position at @cmdi.bsky.social there were no readily available phage against my A. baumannii focal strain. The solution? Finding my own phage ofc. Mystique is a broad host range Acinetobacter phage, and I'm thrilled to see this work out today #PhageSky 🧪🦠
Reposted by Matthew J Shepherd
We're on a mission to #KeepAntibioticsWorking for generations to come, but we need your help
@profkevinfenton.bsky.social, @fphuk.bsky.social, tells us why this is such an important mission
📅 Join us on 7 April to learn exactly what steps you can take to help tackle this growing health challenge
@profkevinfenton.bsky.social, @fphuk.bsky.social, tells us why this is such an important mission
📅 Join us on 7 April to learn exactly what steps you can take to help tackle this growing health challenge
April 3, 2025 at 4:16 PM
We're on a mission to #KeepAntibioticsWorking for generations to come, but we need your help
@profkevinfenton.bsky.social, @fphuk.bsky.social, tells us why this is such an important mission
📅 Join us on 7 April to learn exactly what steps you can take to help tackle this growing health challenge
@profkevinfenton.bsky.social, @fphuk.bsky.social, tells us why this is such an important mission
📅 Join us on 7 April to learn exactly what steps you can take to help tackle this growing health challenge
Reposted by Matthew J Shepherd
Delighted to have won a poster prize at #microbio25 this week! Had a really great time at this conference, learned so much and met so many old (and some new!) faces. Thanks so much to the organisers for a great event and to the society for the travel grant too! 🚝🧪🦠
- Journal of General Virology poster prize winner: Afifah Tasnim @afifahtasnim.bsky.social
-Microbiology poster prize winner: Louise Flanagan @flanagella.bsky.social
-Microbiology poster prize winner: Louise Flanagan @flanagella.bsky.social


April 3, 2025 at 4:57 PM
Delighted to have won a poster prize at #microbio25 this week! Had a really great time at this conference, learned so much and met so many old (and some new!) faces. Thanks so much to the organisers for a great event and to the society for the travel grant too! 🚝🧪🦠
Reposted by Matthew J Shepherd
The MERMan contingent at #microbio25 headed home today after a great week in Liverpool for the @microbiologysociety.org annual conference - Thanks to the society and organisers for putting on a great event, and supporting us in presenting 6 talks and 5 posters packed full of microbial evolution!

April 3, 2025 at 4:11 PM
The MERMan contingent at #microbio25 headed home today after a great week in Liverpool for the @microbiologysociety.org annual conference - Thanks to the society and organisers for putting on a great event, and supporting us in presenting 6 talks and 5 posters packed full of microbial evolution!
Reposted by Matthew J Shepherd
@brockhurstlab.bsky.social giving a talk about eco-evolutionary mechanisms of AMR in the AMR session at #Microbio25

April 1, 2025 at 10:54 AM
@brockhurstlab.bsky.social giving a talk about eco-evolutionary mechanisms of AMR in the AMR session at #Microbio25
Reposted by Matthew J Shepherd
To all enjoying #MicroBio25 remember: all @microbiologysociety.org events rely on our journals - every paper pays 4x travel grants!
Make 2025 when you submit a paper to a MicroSoc journal www.microbiologyresearch.org
$0 OA for Publish+Read institutions www.microbiologyresearch.org/publish-and-...
Make 2025 when you submit a paper to a MicroSoc journal www.microbiologyresearch.org
$0 OA for Publish+Read institutions www.microbiologyresearch.org/publish-and-...

Microbiology Society
Microbiology Society journals contain high-quality research papers and topical review articles. We are a not-for-profit publisher and we support and invest in the microbiology community, to the benefi...
www.microbiologyresearch.org
April 2, 2025 at 7:30 AM
To all enjoying #MicroBio25 remember: all @microbiologysociety.org events rely on our journals - every paper pays 4x travel grants!
Make 2025 when you submit a paper to a MicroSoc journal www.microbiologyresearch.org
$0 OA for Publish+Read institutions www.microbiologyresearch.org/publish-and-...
Make 2025 when you submit a paper to a MicroSoc journal www.microbiologyresearch.org
$0 OA for Publish+Read institutions www.microbiologyresearch.org/publish-and-...
Reposted by Matthew J Shepherd
Had a fab time presenting a flash talk at #microbio25 in the urogenital microbes session this afternoon. If you love a good biofilm or polymicrobial communities and want to find out more, I'd love to chat at my poster tomorrow - B086 👩🏼🏫 @microbiologysociety.org

April 1, 2025 at 6:36 PM
Had a fab time presenting a flash talk at #microbio25 in the urogenital microbes session this afternoon. If you love a good biofilm or polymicrobial communities and want to find out more, I'd love to chat at my poster tomorrow - B086 👩🏼🏫 @microbiologysociety.org
Nice talk by @dralhubb.bsky.social at #microbio25. Urobiome as a possible reservoir of AMR for UTIs, including HGT and selection of spontaneous mutants!

April 1, 2025 at 4:18 PM
Nice talk by @dralhubb.bsky.social at #microbio25. Urobiome as a possible reservoir of AMR for UTIs, including HGT and selection of spontaneous mutants!
Reposted by Matthew J Shepherd
@matthewjshepherd.bsky.social talking about clinical PA in chronic lung infection suggesting picking 10~20 colonies is pretty good for diagnosing pre-existing AMR #Mircobio25

April 1, 2025 at 11:38 AM
@matthewjshepherd.bsky.social talking about clinical PA in chronic lung infection suggesting picking 10~20 colonies is pretty good for diagnosing pre-existing AMR #Mircobio25
Reposted by Matthew J Shepherd
In addition to @brockhurstlab.bsky.social @matthewjshepherd.bsky.social 's upcoming talks in the AMR session today, we have many @mermanchester.bsky.social posters on display! Come learn from our talented scientists about evolution, AMR, and microbiomes #microbio25

April 1, 2025 at 10:19 AM
In addition to @brockhurstlab.bsky.social @matthewjshepherd.bsky.social 's upcoming talks in the AMR session today, we have many @mermanchester.bsky.social posters on display! Come learn from our talented scientists about evolution, AMR, and microbiomes #microbio25