@gladstoneinst.bsky.social @czbiohub.bsky.social
👉Human variant interpretation with sequence-to-activity models
📅Wed June 4,5:30pm CET🧬 kipoi.org/seminar/🦋@kipoizoo.bsky.social
@gladstoneinst.bsky.social @czbiohub.bsky.social
👉Human variant interpretation with sequence-to-activity models
📅Wed June 4,5:30pm CET🧬 kipoi.org/seminar/🦋@kipoizoo.bsky.social
🐕scooby: Modeling multi-modal genomic profiles from DNA sequence at single-cell resolution
📅Wed May 7, 5:30pm CET
🧬https://kipoi.org/seminar/
🦋kipoizoo.bsky
🐕scooby: Modeling multi-modal genomic profiles from DNA sequence at single-cell resolution
📅Wed May 7, 5:30pm CET
🧬https://kipoi.org/seminar/
🦋kipoizoo.bsky

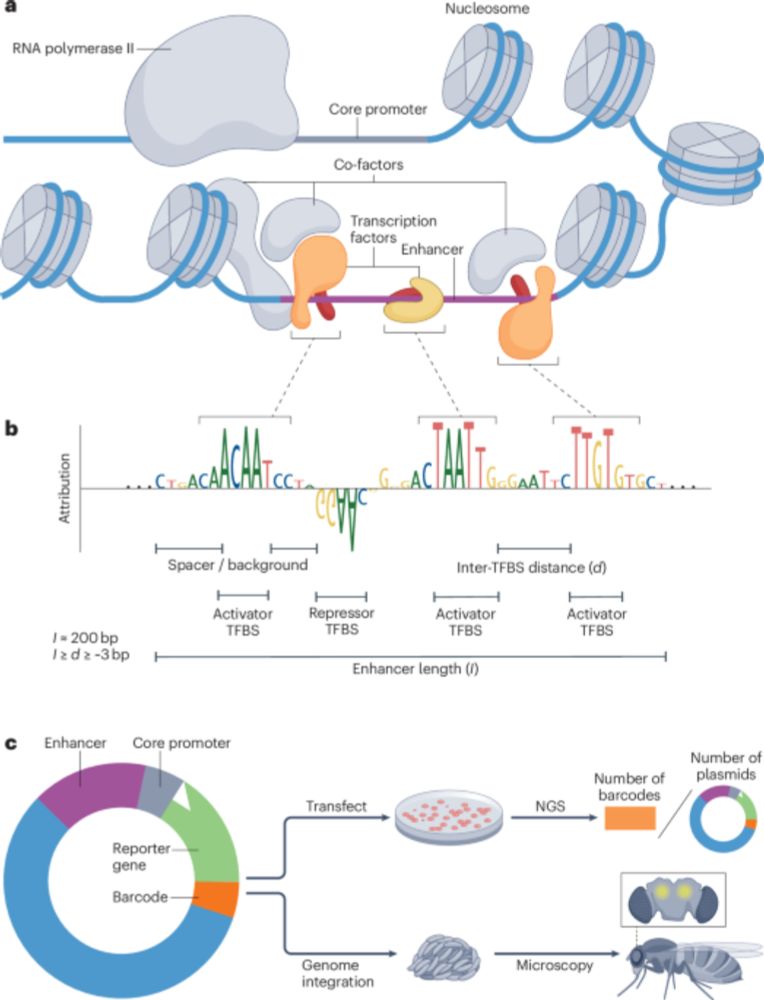
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
doi.org/10.1101/2024...

doi.org/10.1101/2024...

@lxsasse.bsky.social
@zmbh.uni-heidelberg.de
👉Advanced training strategies for genomic sequence-to-function models
📅 Wed March 5, 5:30pm CET
🧬 kipoi.org/seminar/
🦋 @kipoizoo.bsky.social
@lxsasse.bsky.social
@zmbh.uni-heidelberg.de
👉Advanced training strategies for genomic sequence-to-function models
📅 Wed March 5, 5:30pm CET
🧬 kipoi.org/seminar/
🦋 @kipoizoo.bsky.social
👉Nucleotide dependency analysis of DNA language models reveals genomic functional elements
📅Wed Feb 5, 5:30pm CET
🧬https://kipoi.org/seminar/
🦋kipoizoo.bsky
👉Nucleotide dependency analysis of DNA language models reveals genomic functional elements
📅Wed Feb 5, 5:30pm CET
🧬https://kipoi.org/seminar/
🦋kipoizoo.bsky
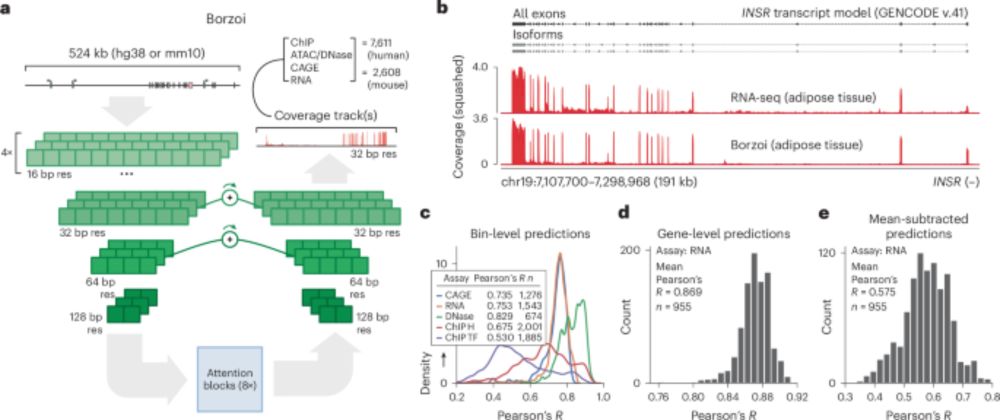
www.nature.com/articles/s41... 1/
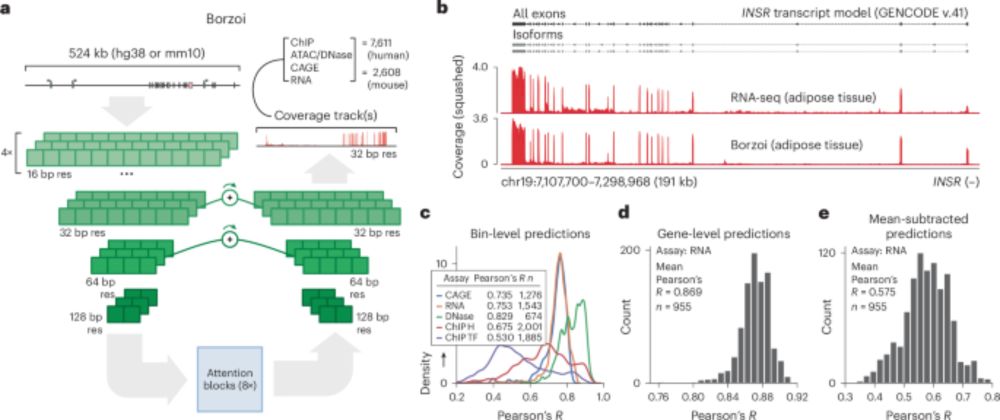
www.nature.com/articles/s41... 1/

This was an incredible team effort w/ @jennifergarrison.bsky.social #TammyLan @davidsebfischer.bsky.social #AlisonKochersberger #RuthRaichur #SophiaSzady
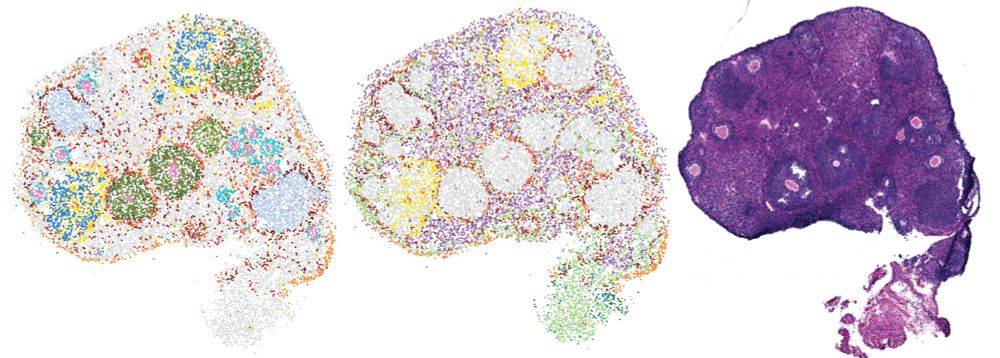
This was an incredible team effort w/ @jennifergarrison.bsky.social #TammyLan @davidsebfischer.bsky.social #AlisonKochersberger #RuthRaichur #SophiaSzady

Join us for our next Kipoi Seminar with with Dmitry Penzar,
@pensarata.bsky.social @ autosome.org!
👉LegNet: parameter-efficient modeling of gene regulatory regions using modern convolutional neural network
📅Wed Dec 4, 5:30pm CET
🧬 kipoi.org/seminar/
Join us for our next Kipoi Seminar with with Dmitry Penzar,
@pensarata.bsky.social @ autosome.org!
👉LegNet: parameter-efficient modeling of gene regulatory regions using modern convolutional neural network
📅Wed Dec 4, 5:30pm CET
🧬 kipoi.org/seminar/
So how can we ever achieve #gender #equality?
We discuss various strategies in this #Cell Commentary
#WomeninSTEM
www.cell.com/cell/fulltex...
🧵 1/
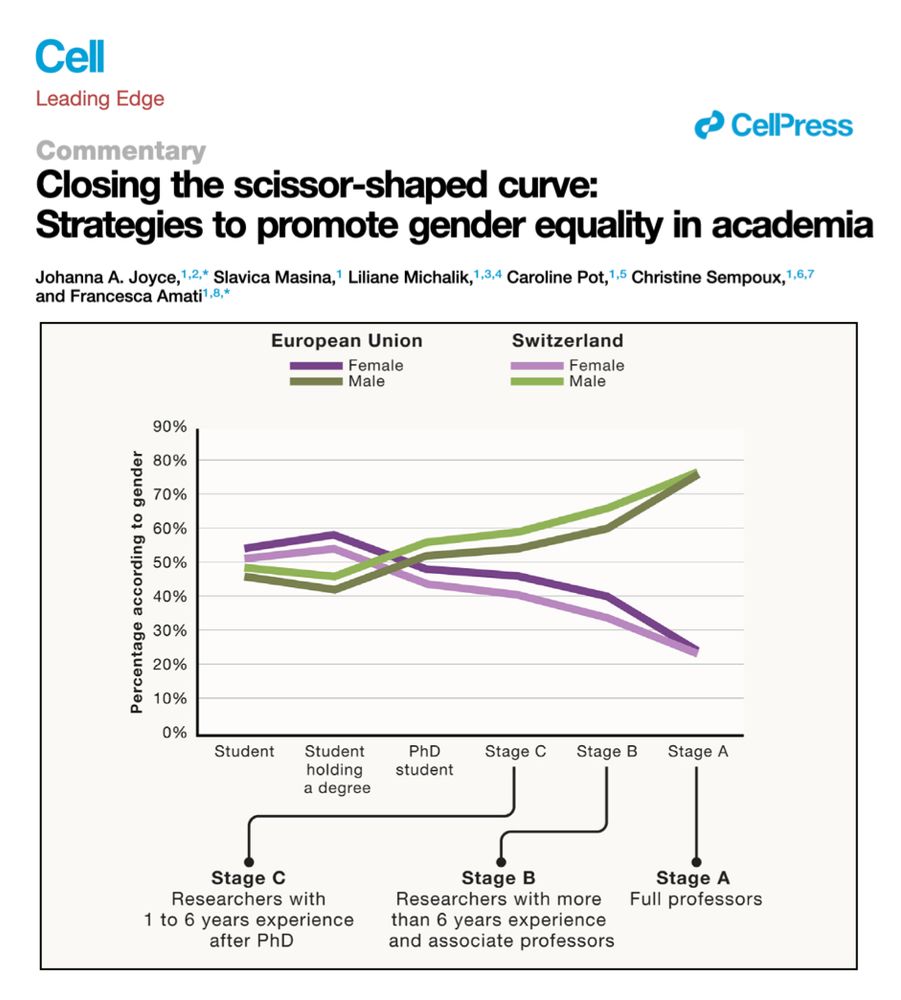
So how can we ever achieve #gender #equality?
We discuss various strategies in this #Cell Commentary
#WomeninSTEM
www.cell.com/cell/fulltex...
🧵 1/
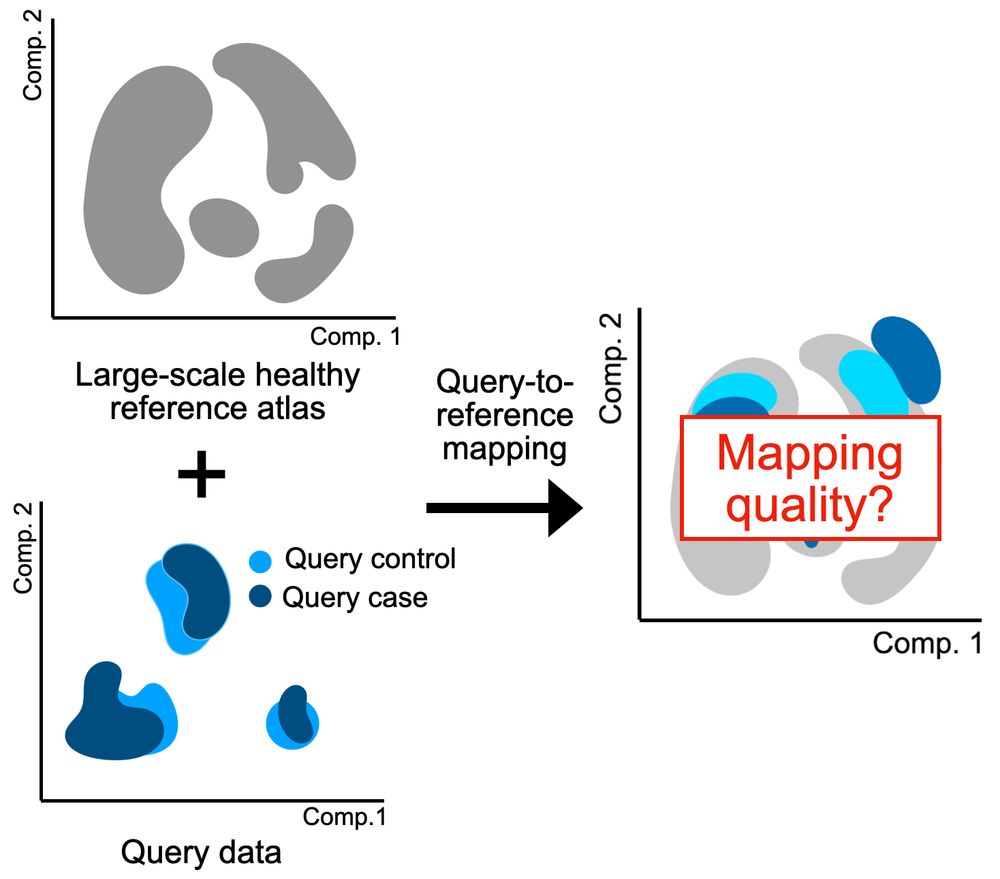
Find out how it can serve you ⏬
🧵1/8
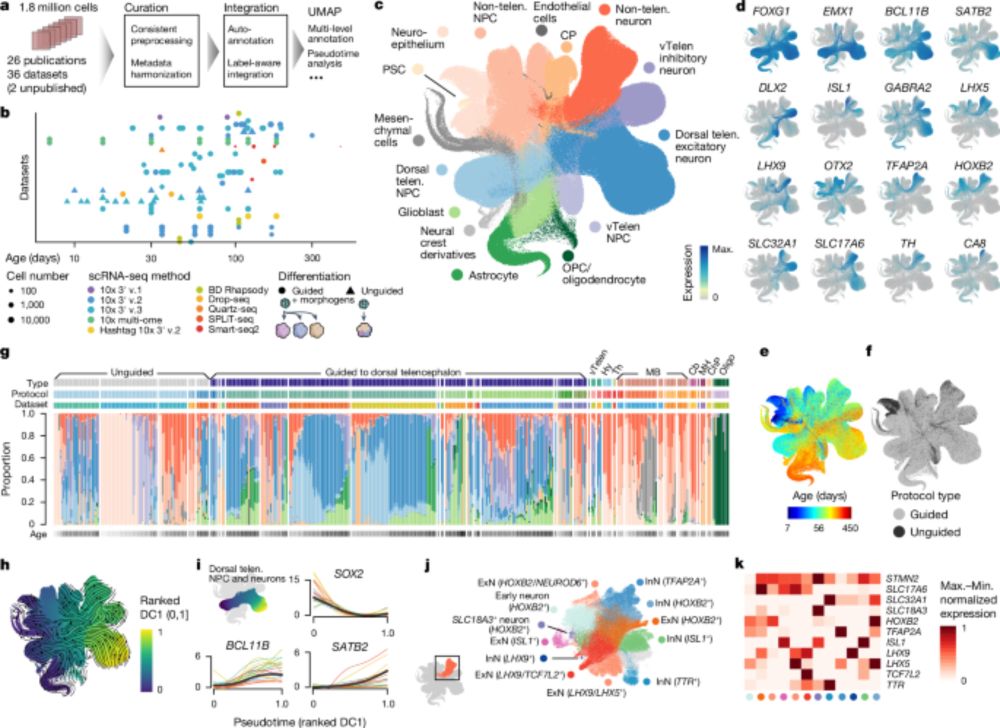
Find out how it can serve you ⏬
🧵1/8
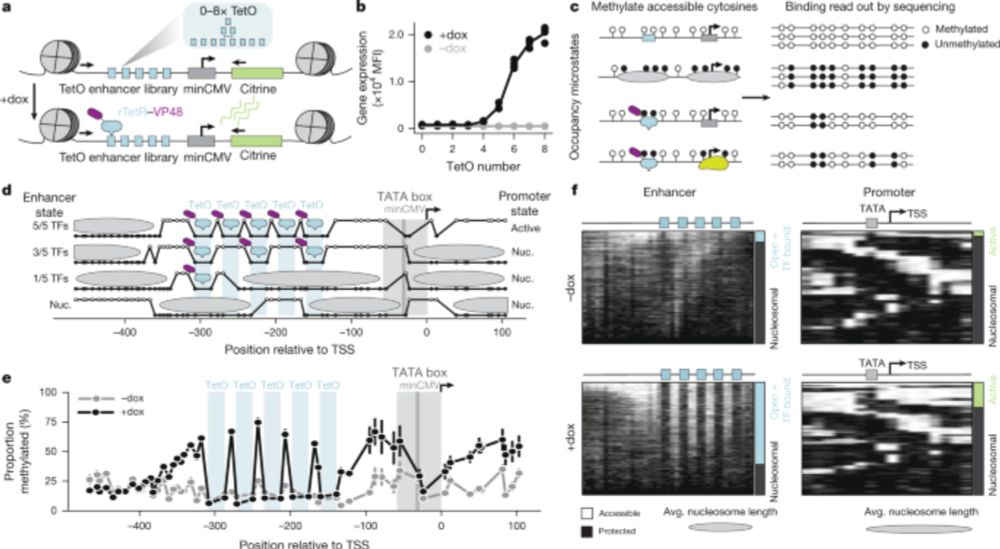
www.nature.com/articles/s41...
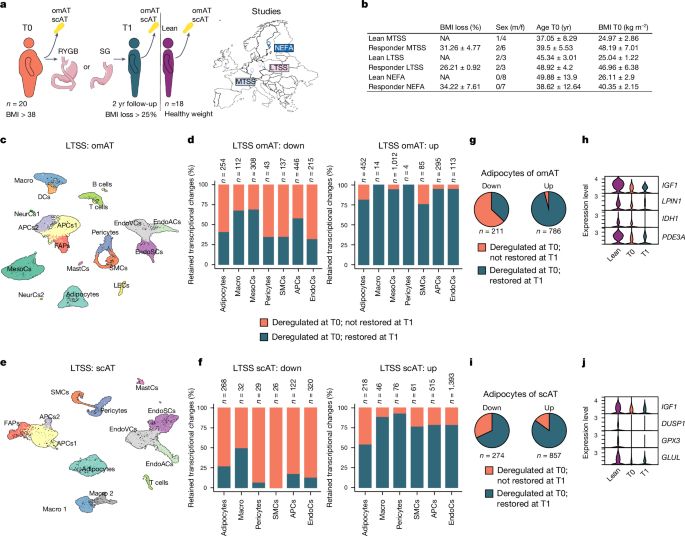
www.nature.com/articles/s41...
If you are interested in doing a Ph.D. with me at UMass Chan Medical (Genomics and Comp Bio Department), see the links below. Deadline is Dec 1st.
If you are interested in doing a Ph.D. with me at UMass Chan Medical (Genomics and Comp Bio Department), see the links below. Deadline is Dec 1st.


