kabylda.github.io
Explore the research behind the art 🔗: buff.ly/J3R4JpL
#ChemSky

Explore the research behind the art 🔗: buff.ly/J3R4JpL
#ChemSky

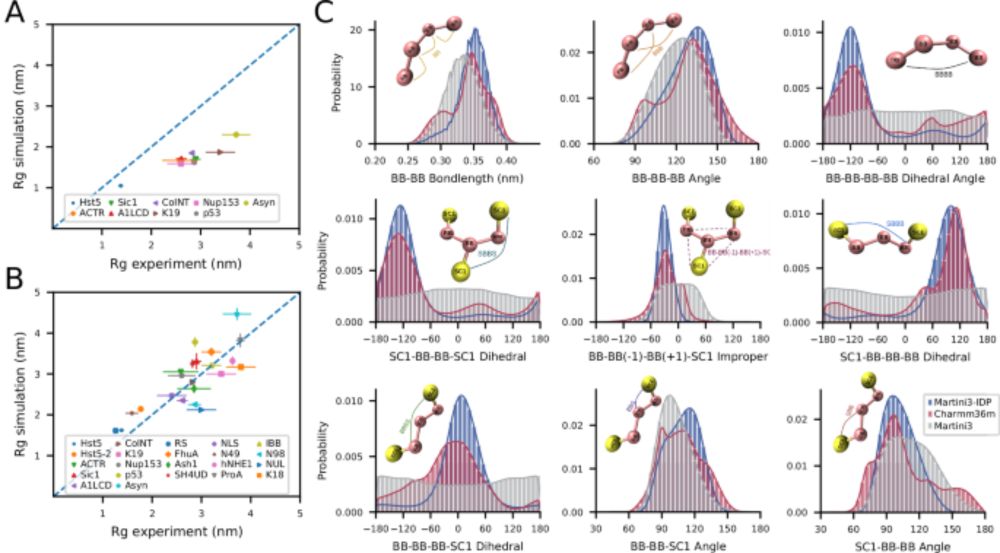
arxiv.org/abs/2503.19847
🧵⬇️

arxiv.org/abs/2503.19847
🧵⬇️
doi.org/10.1038/s415...
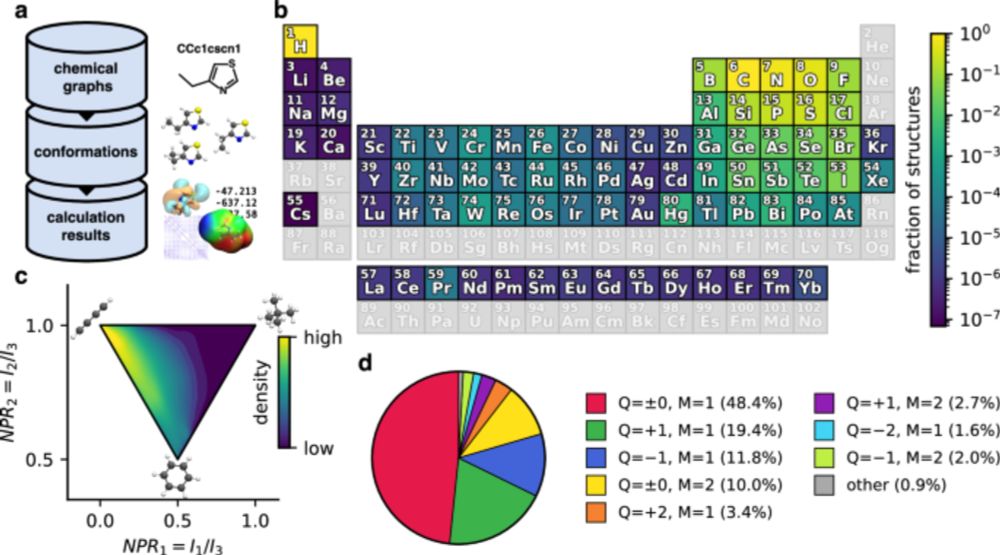
doi.org/10.1038/s415...
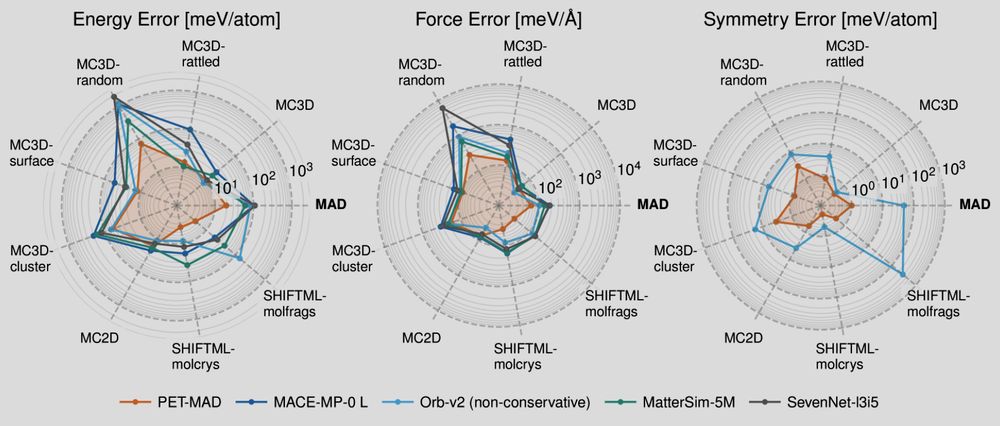
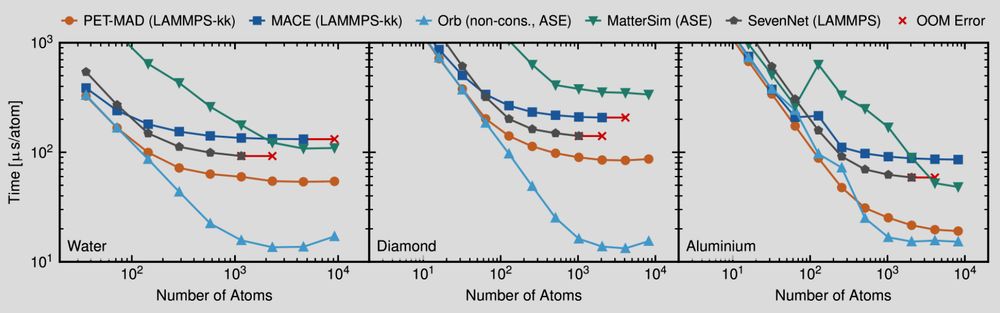

doi.org/10.1021/acs....

doi.org/10.1021/acs....
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
🗓 Deadline: 31/01/25
📥 Application: shorturl.at/nHyAo

🗓 Deadline: 31/01/25
📥 Application: shorturl.at/nHyAo
The representations describe the distance and orientation between atoms, crucial for modeling molecular systems
tinyurl.com/47xud8nr
#MachineLearning #AI4Science
The representations describe the distance and orientation between atoms, crucial for modeling molecular systems
tinyurl.com/47xud8nr
#MachineLearning #AI4Science
SPINACH ON THE CEILING: A Theoretical Chemist's Return to Biology by Martin Karplus. #NMRchat #chemsky
www.annualreviews.org/content/jour...
SPINACH ON THE CEILING: A Theoretical Chemist's Return to Biology by Martin Karplus. #NMRchat #chemsky
www.annualreviews.org/content/jour...

