Nanopore Sequencing Lab
Institute of Medical Genetics
Medical University of Vienna


academic.oup.com/gigascience/...

academic.oup.com/gigascience/...

How about the matching coordinates of an assembly mapped to a reference but repeats and SVs are confusing liftover?
Well I did while trying to benchmark STR stuff. Nice side quest.
github.com/Psy-Fer/bedp...

How about the matching coordinates of an assembly mapped to a reference but repeats and SVs are confusing liftover?
Well I did while trying to benchmark STR stuff. Nice side quest.
github.com/Psy-Fer/bedp...
Try
options(renv.config.pak.enabled = TRUE)
which will use pak to download and install packages *in parallel*. So much faster.
Try
options(renv.config.pak.enabled = TRUE)
which will use pak to download and install packages *in parallel*. So much faster.
shinyelectron::export() → #rshinylive conversion → .dmg → Native Mac app
Zero #rstats dependencies for end users! Early days but promising 👀
shinyelectron::export() → #rshinylive conversion → .dmg → Native Mac app
Zero #rstats dependencies for end users! Early days but promising 👀
The link you gave is good but points to a 2yr old rendering. This one uses recent versions of R and all packages and has all the latest typo fixes:
www.huber.embl.de/msmb/

The link you gave is good but points to a 2yr old rendering. This one uses recent versions of R and all packages and has all the latest typo fixes:
www.huber.embl.de/msmb/
Learn more: youtu.be/FL8ZKUmtYbM
#EurekaPrizes

@bosc.bsky.social track around 2:30pm ish after
@sunethsa.bsky.social's talk, Bonson Wong will present on
nanopore basecalling on AMD GPUs using slorado
github.com/BonsonW/slor...
@bosc.bsky.social track around 2:30pm ish after
@sunethsa.bsky.social's talk, Bonson Wong will present on
nanopore basecalling on AMD GPUs using slorado
github.com/BonsonW/slor...

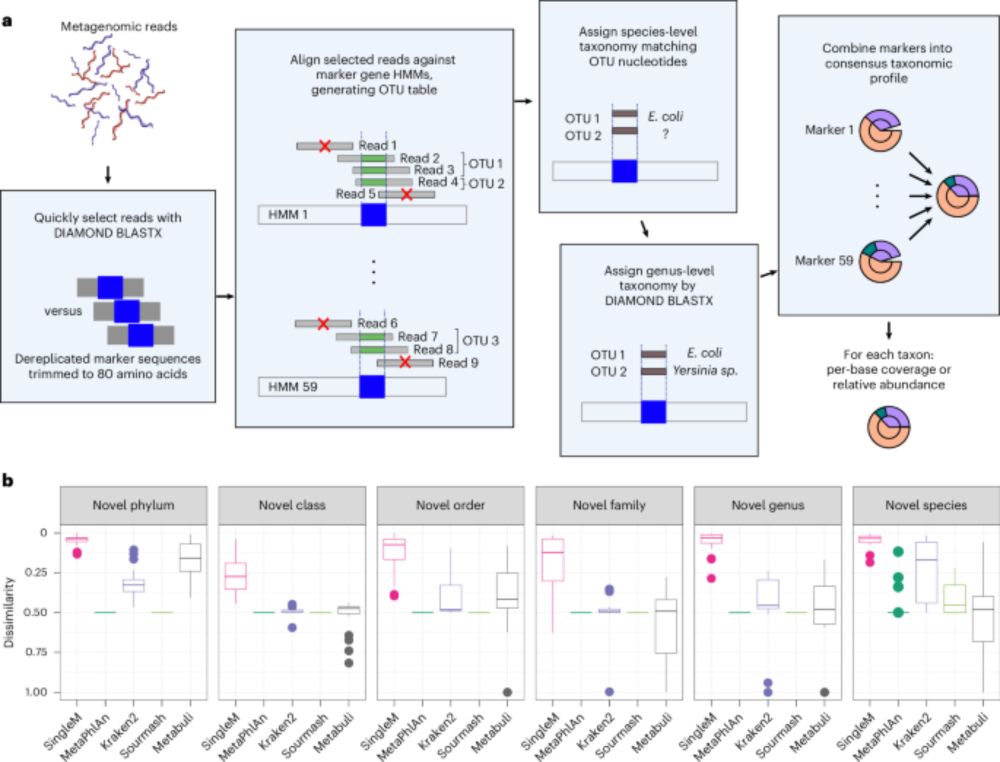
It’s faster, more accurate, and ready for thousands of genomes
Let’s break it down (1/10)
github.com/OrthoFinder/...
www.biorxiv.org/content/10.1...
It’s faster, more accurate, and ready for thousands of genomes
Let’s break it down (1/10)
github.com/OrthoFinder/...
www.biorxiv.org/content/10.1...
Learn more: posit.co/blog/introdu...

github.com/warp9seq/min...
written by
@sunethsa.bsky.social
in C, based on mod tag parsing we did for realfreq doi.org/10.1093/bioi...
github.com/warp9seq/min...
written by
@sunethsa.bsky.social
in C, based on mod tag parsing we did for realfreq doi.org/10.1093/bioi...
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
biorxiv.org/content/10.1...
Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
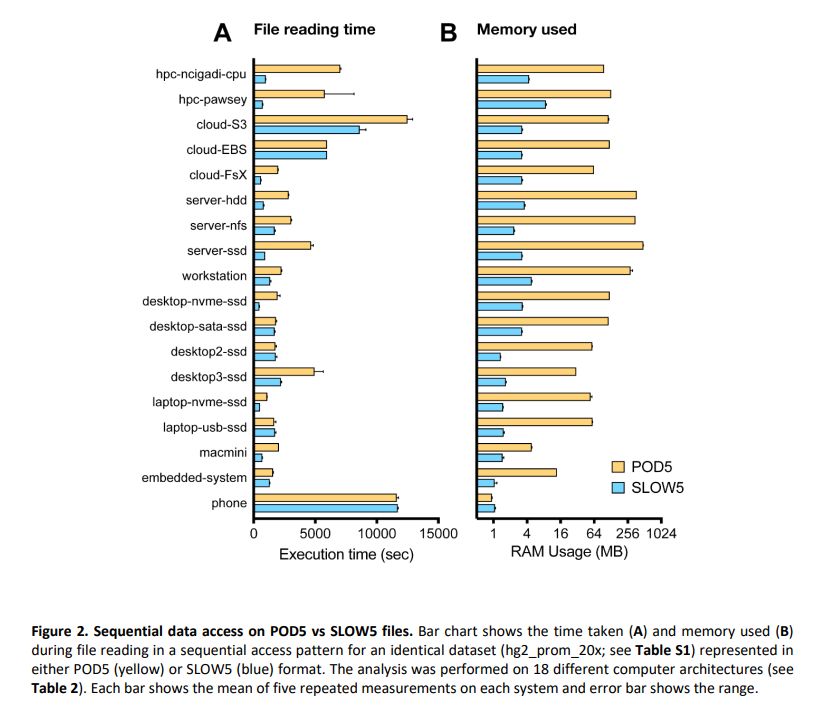
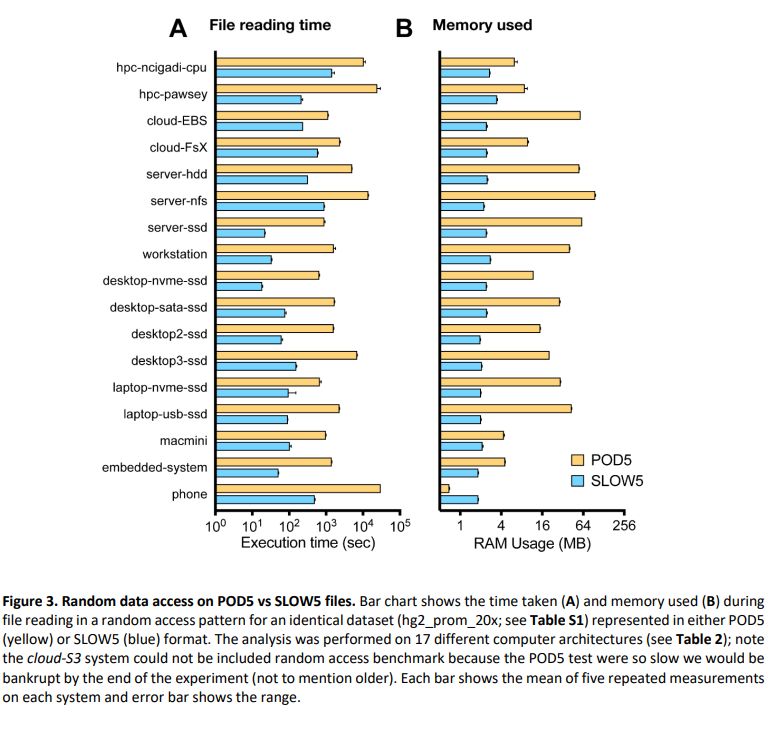
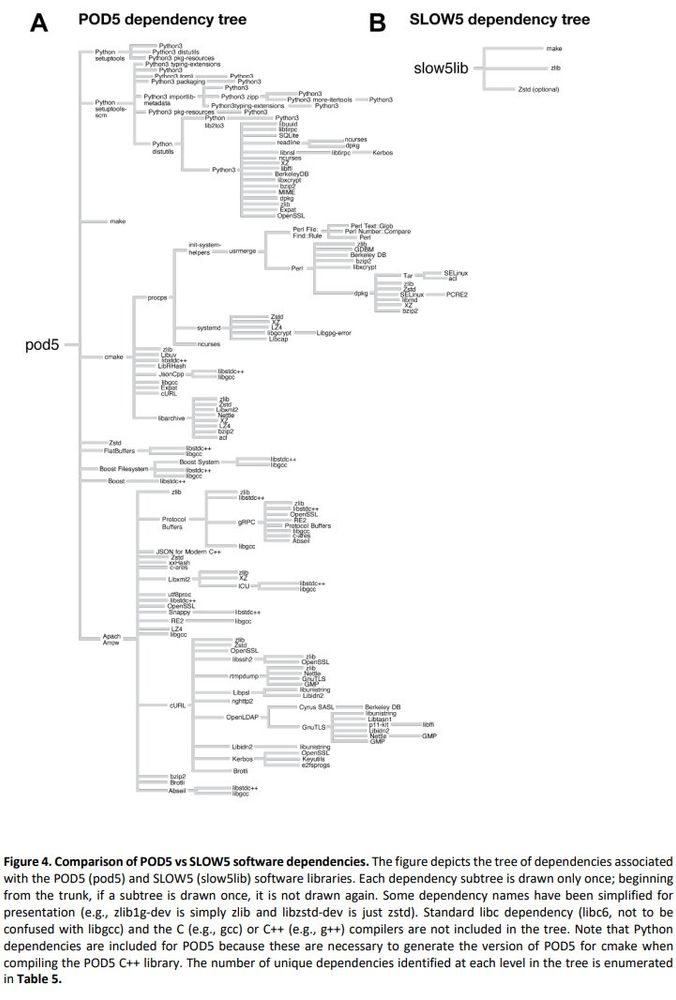
biorxiv.org/content/10.1...
Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
https://ziglang.org/devlog/2025/#2025-06-14
https://ziglang.org/devlog/2025/#2025-06-14
myloasm-docs.github.io
myloasm-docs.github.io


- greatly reduces cost per genome assembly
- reference agnostic, so works for non-humans
- assembly just using saliva
- & many more
Relies on 2 excellent software #readfish & #hifiasm.
- greatly reduces cost per genome assembly
- reference agnostic, so works for non-humans
- assembly just using saliva
- & many more
Relies on 2 excellent software #readfish & #hifiasm.