I'm mostly interested in human history and evolution, but also more general: Bioinformatics, Population genetics, Phylogenetics, aDNA, Simulation tools and Palaeoproteomics.
@ipatramanis.bsky.social PAASTA talk from March about PaleoProPhyler a tool for #phylogenies from ancient proteins is now live on our YouTube youtu.be/2gVEQyinL4A

@ipatramanis.bsky.social PAASTA talk from March about PaleoProPhyler a tool for #phylogenies from ancient proteins is now live on our YouTube youtu.be/2gVEQyinL4A
Than you are more than welcome to apply to join my group starting Jan 2026 :)
candidate.hr-manager.net/ApplicationI...
Please reach out if you have any questions!

Than you are more than welcome to apply to join my group starting Jan 2026 :)
candidate.hr-manager.net/ApplicationI...
Please reach out if you have any questions!
go.nature.com/4lFx4KN

go.nature.com/4lFx4KN
www.greenmatters.com/news/lynch-s...

www.greenmatters.com/news/lynch-s...
See the post from Palesa for a quick summary.
Although technically small in scale (4 specimens🦷), this work took many years and many people to complete...
Excited to see how the results we show will be applied to future sample sets, now that we know what works!
Our paper on enamel proteins from Paranthropus robustus has finally been peer reviewed, please have a read here: www.science.org/doi/10.1126/...
Paranthropus robustus has been puzzling scientists since its discovery in 1938 in South Africa, where a high number of fossils have been found.
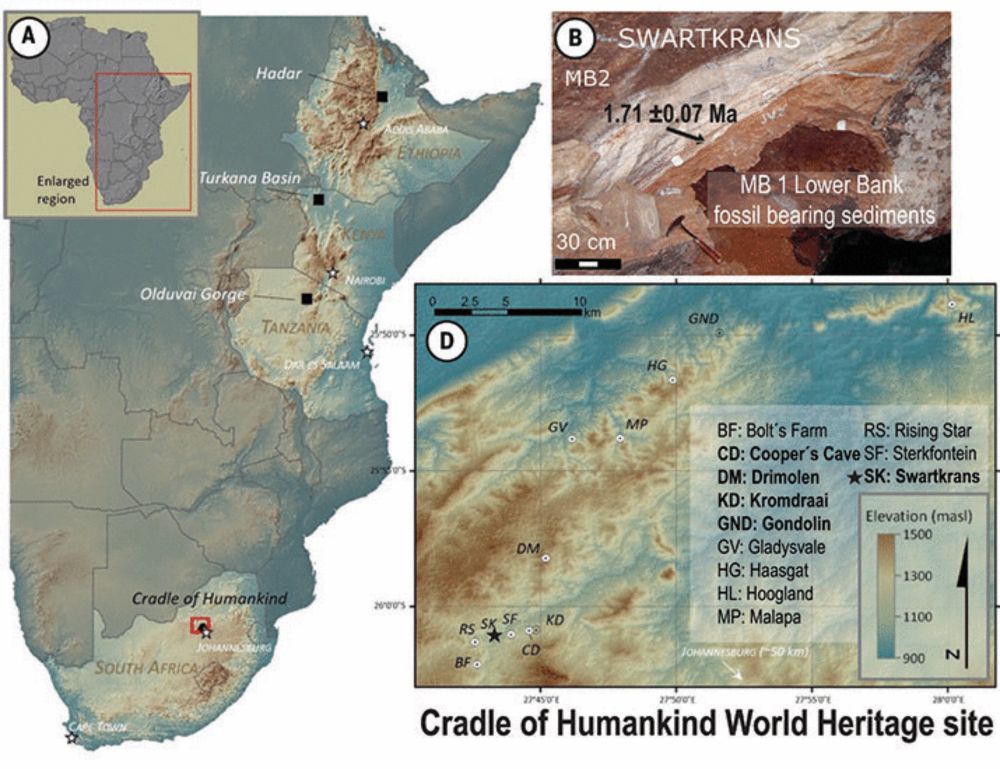
See the post from Palesa for a quick summary.
Although technically small in scale (4 specimens🦷), this work took many years and many people to complete...
Excited to see how the results we show will be applied to future sample sets, now that we know what works!
This work has been led by @ebosch1972.bsky.social @francesccalafell.bsky.social ( @upf.edu ) in collaboration with Rafael de Cid ( @igtp.bsky.social ) and @sabiagini.bsky.social
⬇️⬇️⬇️
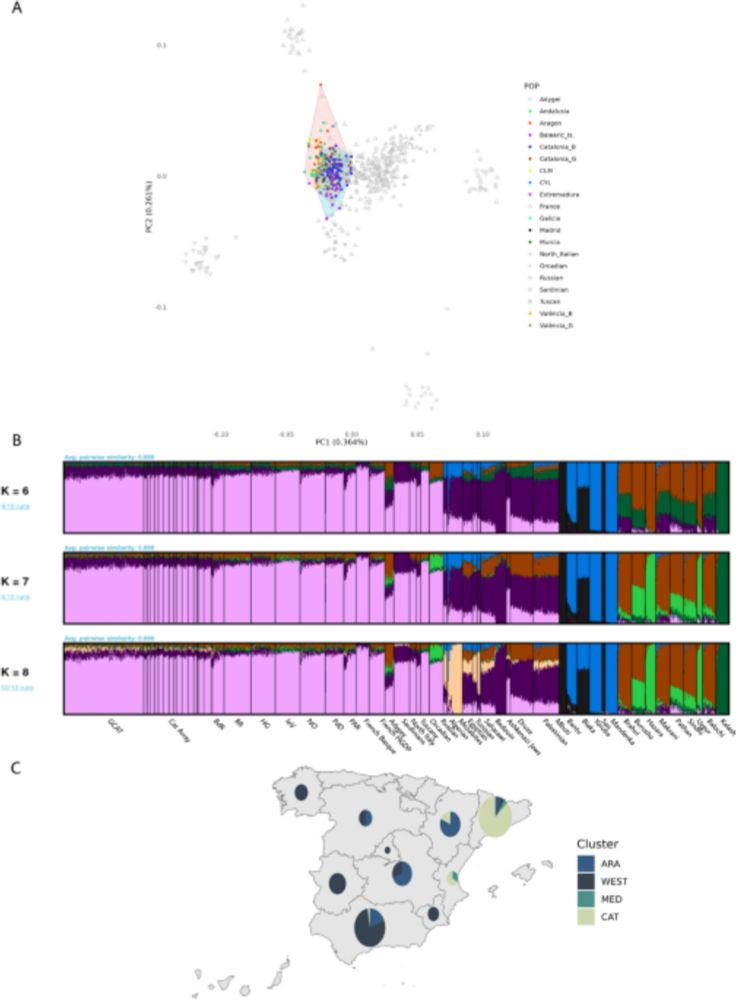
This work has been led by @ebosch1972.bsky.social @francesccalafell.bsky.social ( @upf.edu ) in collaboration with Rafael de Cid ( @igtp.bsky.social ) and @sabiagini.bsky.social
⬇️⬇️⬇️
Over the years, I’ve gotten multiple questions on the usefulness of ancient proteins in resolving phylogenetic questions. This is our attempt to provide some answers:
www.biorxiv.org/content/10.1...

Over the years, I’ve gotten multiple questions on the usefulness of ancient proteins in resolving phylogenetic questions. This is our attempt to provide some answers:
www.biorxiv.org/content/10.1...
@science.org! doi.org/10.1126/scie...
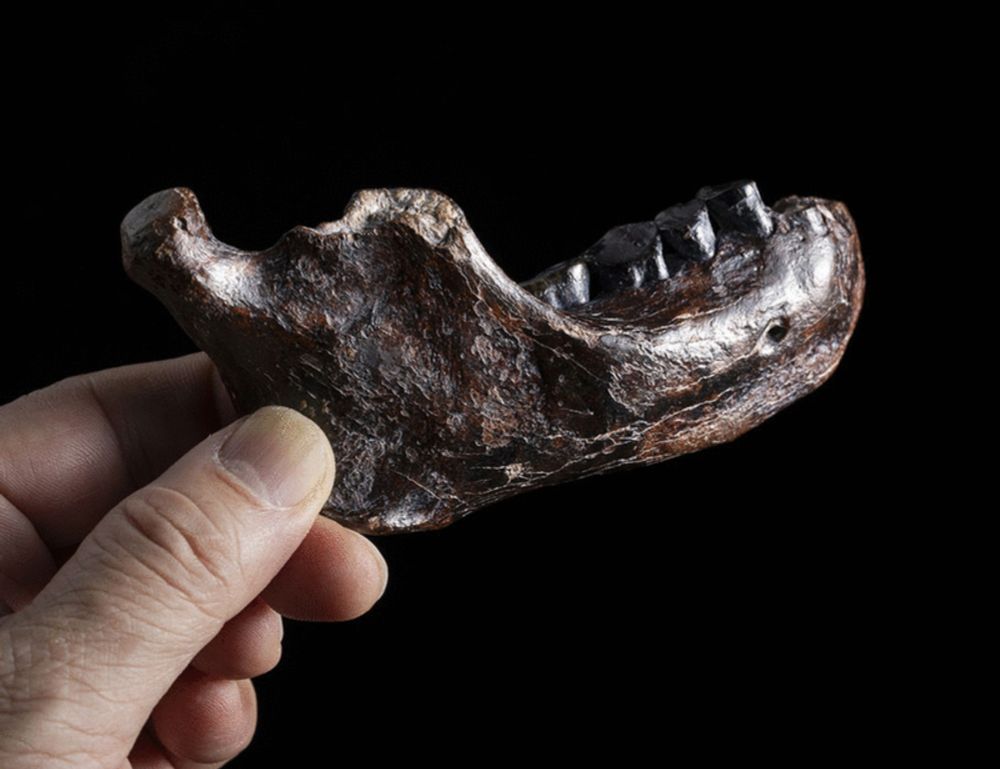
@science.org! doi.org/10.1126/scie...
We contributed towards the South African Journal of Science Taung Centennial!
some links;
the special issue: issuu.com/sajs/docs/so...
our paper: sajs.co.za/article/view...
and I got featured on where I work from Nature Africa if you are interested in that: www.nature.com/articles/d44...


We contributed towards the South African Journal of Science Taung Centennial!
some links;
the special issue: issuu.com/sajs/docs/so...
our paper: sajs.co.za/article/view...
and I got featured on where I work from Nature Africa if you are interested in that: www.nature.com/articles/d44...
Paleoproteomics sheds light on million-year-old fossils 🏺🧪
www.nature.com/articles/s41...

Paleoproteomics sheds light on million-year-old fossils 🏺🧪
www.nature.com/articles/s41...

