
@UBC
. Keeping it 100K.
pubmed.ncbi.nlm.nih.gov/41123210/ #cryoEM ❄️🔬🇨🇦

pubmed.ncbi.nlm.nih.gov/41123210/ #cryoEM ❄️🔬🇨🇦
#cryoEM #structuralbiology
www.nature.com/articles/s41...
#cryoEM #structuralbiology
www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Congratulations to Valentina Francia, Yao Zhang and colleagues from the Cullis lab at UBC.
We are proud to have supported this work! 🔬
doi.org/10.1073/pnas...

Congratulations to Valentina Francia, Yao Zhang and colleagues from the Cullis lab at UBC.
We are proud to have supported this work! 🔬
doi.org/10.1073/pnas...
Topic: CryoDRGN-AI: Neural ab initio reconstruction of challenging cryo-EM and cryo-ET datasets
Host: Pete Meyer
Recorded on: May 20, 2025
buff.ly/jKjWTOa
#SBGridWebinars

Topic: CryoDRGN-AI: Neural ab initio reconstruction of challenging cryo-EM and cryo-ET datasets
Host: Pete Meyer
Recorded on: May 20, 2025
buff.ly/jKjWTOa
#SBGridWebinars
Shout to Katrien Willegems, Jodene Eldstrom, Efthimios Kyriakis, Fariba Ataei and researchers from the Fedida and Van Petegem Labs at UBC! 🔬🫀🧬
#cryoEM
www.nature.com/articles/s41...

Shout to Katrien Willegems, Jodene Eldstrom, Efthimios Kyriakis, Fariba Ataei and researchers from the Fedida and Van Petegem Labs at UBC! 🔬🫀🧬
#cryoEM
www.nature.com/articles/s41...
We are proud to have provided the electron microscopy for this work!
Read more:
doi.org/10.1016/j.st...

We are proud to have provided the electron microscopy for this work!
Read more:
doi.org/10.1016/j.st...
New research from @coonlab.bsky.social and the Grant Lab combines mass spec to land proteins on TEM grids, a laser to restore protein structure, and cryoEM imaging and analysis to produce 3D reconstructions.
www.mcponline.org/article/S153...

New research from @coonlab.bsky.social and the Grant Lab combines mass spec to land proteins on TEM grids, a laser to restore protein structure, and cryoEM imaging and analysis to produce 3D reconstructions.
www.mcponline.org/article/S153...
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
We are happy to have supported this work! #cryoEM
Read more:
www.nature.com/articles/s41...

We are happy to have supported this work! #cryoEM
Read more:
www.nature.com/articles/s41...
Shout out to @elitzatocheva.bsky.social !
Read more about this awesome work supported by HRMEM here:
www.pnas.org/doi/10.1073/...

Shout out to @elitzatocheva.bsky.social !
Read more about this awesome work supported by HRMEM here:
www.pnas.org/doi/10.1073/...
We expanded Surface Morphometrics to quantify membrane thickness from cryo-ET—revealing local variation across organelles.
Led by the lab’s first grad student, @mmedina300kv.bsky.social (defending Monday! 🍾) w/ @attychang.bsky.social @hamid13r.bsky.social & @tomo.science
We expanded Surface Morphometrics to quantify membrane thickness from cryo-ET—revealing local variation across organelles.
Led by the lab’s first grad student, @mmedina300kv.bsky.social (defending Monday! 🍾) w/ @attychang.bsky.social @hamid13r.bsky.social & @tomo.science
HRMEM is happy to have supported the electron microscopy for this exciting research!
Read more :
doi.org/10.1016/j.bi...

HRMEM is happy to have supported the electron microscopy for this exciting research!
Read more :
doi.org/10.1016/j.bi...
@berkeleylab.lbl.gov 's Peter Denes, "but it should enable microscopists to image even proteins roughly half the size of what is currently possible."
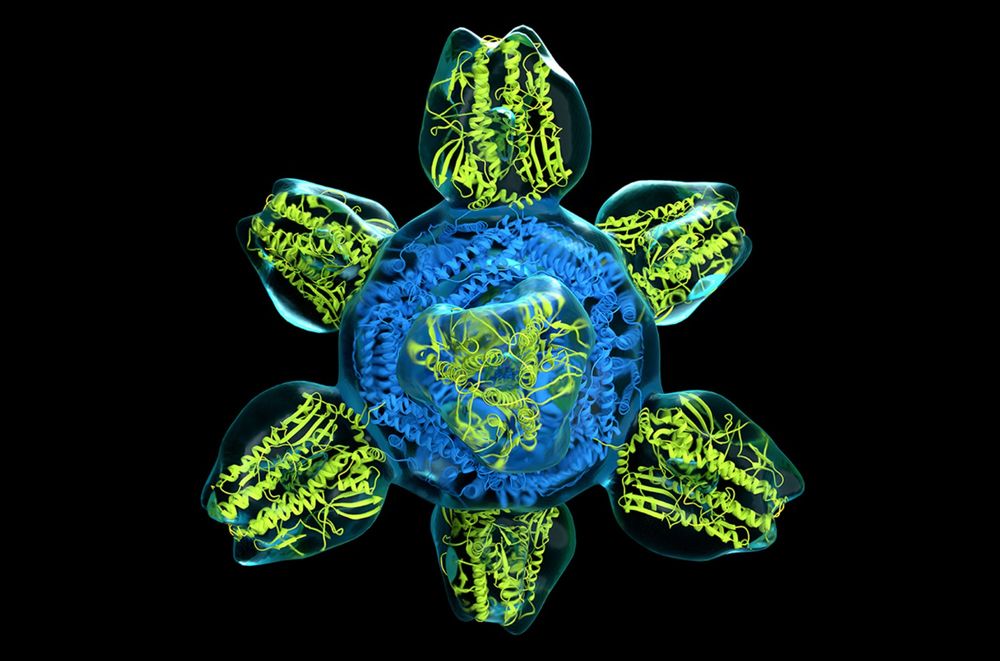
@berkeleylab.lbl.gov 's Peter Denes, "but it should enable microscopists to image even proteins roughly half the size of what is currently possible."
It was a pleasure to collaborate with the Van Petegem (@filipvanpetegem.bsky.social) Lab on this lovely work!
www.nature.com/articles/s41...

It was a pleasure to collaborate with the Van Petegem (@filipvanpetegem.bsky.social) Lab on this lovely work!
www.nature.com/articles/s41...
Congratulations to the Burke Lab (@jburkevic.bsky.social )at the University of Victoria and the Yip Lab at UBC! 🔬🎉
Read more here:
www.nature.com/articles/s41...

Congratulations to the Burke Lab (@jburkevic.bsky.social )at the University of Victoria and the Yip Lab at UBC! 🔬🎉
Read more here:
www.nature.com/articles/s41...
HRMEM is proud to have supported this work! 🔬🫶
www.nature.com/articles/s41...

HRMEM is proud to have supported this work! 🔬🫶
www.nature.com/articles/s41...
Read more in Nature: www.nature.com/articles/s41... 🧪
🧵 on methods & TB biology...
www.pnas.org/doi/full/10....
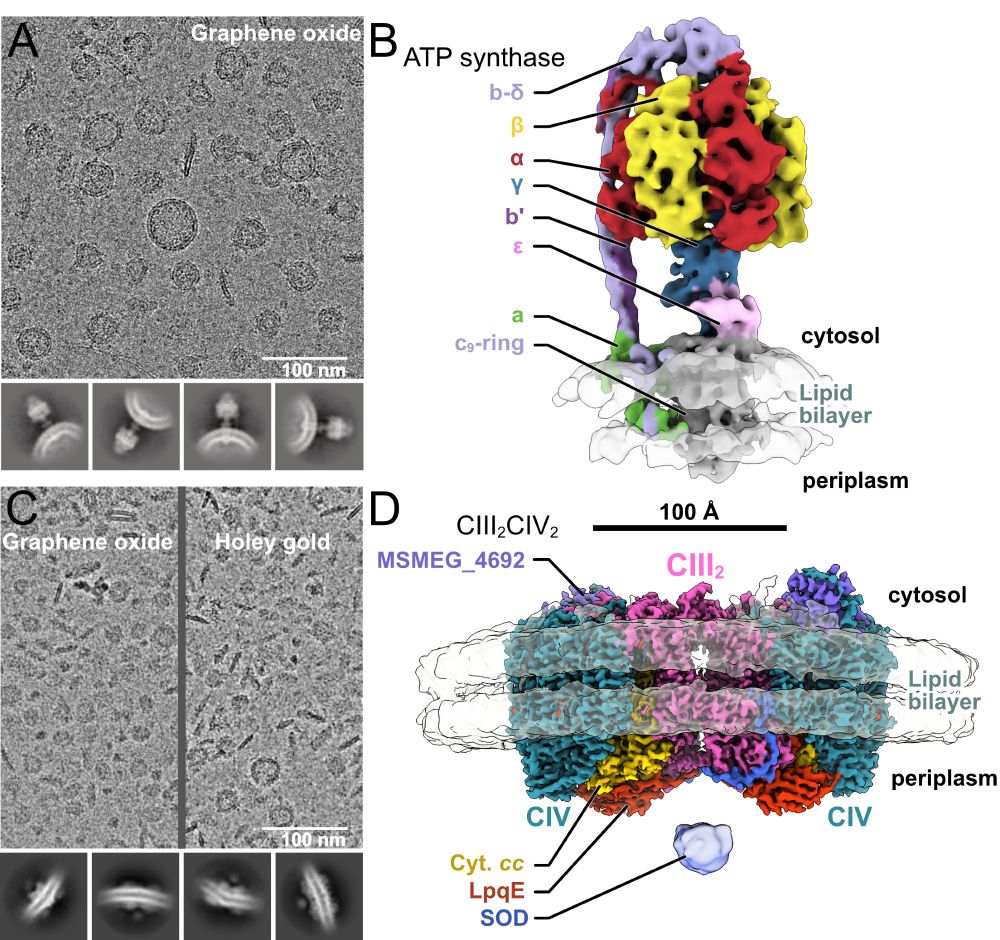
🧵 on methods & TB biology...
www.pnas.org/doi/full/10....
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


