
looking to hire 2 postdocs to join us in this endeavor.
more details can be found here: sites.google.com/uw.edu/statp...
looking to hire 2 postdocs to join us in this endeavor.
more details can be found here: sites.google.com/uw.edu/statp...
Can you predict immune state labels from adaptive immune receptor repertoires?
💰 $10,000 prize pool!
🗓️ Launches Nov 5 on @kaggle.com
More Info: uio-bmi.github.io/adaptive_imm...

Can you predict immune state labels from adaptive immune receptor repertoires?
💰 $10,000 prize pool!
🗓️ Launches Nov 5 on @kaggle.com
More Info: uio-bmi.github.io/adaptive_imm...
We were motivated to fit wide-context mutation models based on previous analyses showing "mesoscale" effects and a position-specific effect. But, how to avoid exploding the number of parameters? 🧵

We were motivated to fit wide-context mutation models based on previous analyses showing "mesoscale" effects and a position-specific effect. But, how to avoid exploding the number of parameters? 🧵

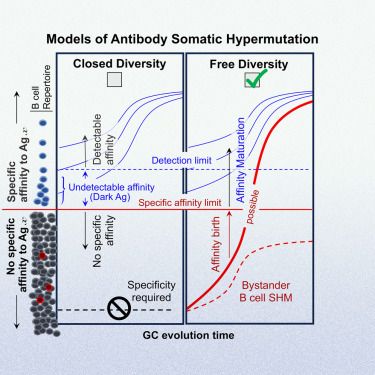
academic.oup.com/mbe/advance-...
Are you curious about how fast the genome of E.coli evolves structurally (gains, rearrangements...) ? 🧬
A summary thread [1/N]🧵

academic.oup.com/mbe/advance-...
Are you curious about how fast the genome of E.coli evolves structurally (gains, rearrangements...) ? 🧬
A summary thread [1/N]🧵

💻🧬TCR-TRANSLATE - A new framework for thinking about the TCR:pMHC specificity problem.
TLDR:
We pretrained LLMs on ~8M TCR & pMHC seqs
Finetuned on sparse pMHC->TCR pair data
Validated CDR3b sequences to unseen antigens
>> random performance on IMMREP2023 "private" antigens

💻🧬TCR-TRANSLATE - A new framework for thinking about the TCR:pMHC specificity problem.
TLDR:
We pretrained LLMs on ~8M TCR & pMHC seqs
Finetuned on sparse pMHC->TCR pair data
Validated CDR3b sequences to unseen antigens
>> random performance on IMMREP2023 "private" antigens

