(Seattle)
looking to hire 2 postdocs to join us in this endeavor.
more details can be found here: sites.google.com/uw.edu/statp...
looking to hire 2 postdocs to join us in this endeavor.
more details can be found here: sites.google.com/uw.edu/statp...
Apply: apply.interfolio.com/172697
Faculty: www.fredhutch.org/en/research...

Apply: apply.interfolio.com/172697
Faculty: www.fredhutch.org/en/research...
www.pnas.org/doi/10.1073/...
Led by Gian Marco Visani (effort initiated by Michael Pun), fantastic collaboration with @pgtimmune.bsky.social @asya-minervina.bsky.social and Phil Bradley.
www.pnas.org/doi/10.1073/...
Led by Gian Marco Visani (effort initiated by Michael Pun), fantastic collaboration with @pgtimmune.bsky.social @asya-minervina.bsky.social and Phil Bradley.

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


structure-based machine learning model for TCR-pMHC complexes, predicting T-cell affinity to peptide-MHC complexes, quantifying T-cell receptor specificity, and designing de-novo immunogenic peptides:
arxiv.org/abs/2503.00648
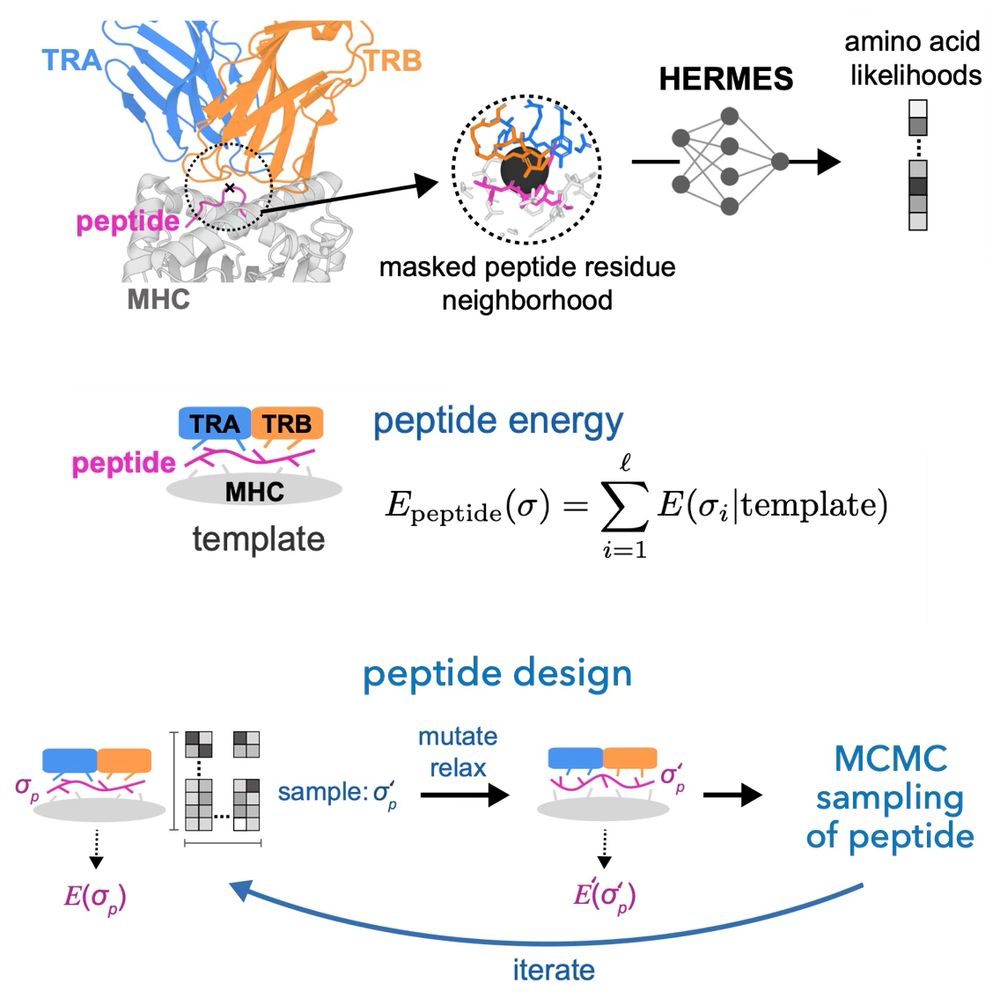
structure-based machine learning model for TCR-pMHC complexes, predicting T-cell affinity to peptide-MHC complexes, quantifying T-cell receptor specificity, and designing de-novo immunogenic peptides:
arxiv.org/abs/2503.00648

