
Left X-Twitter on Nov. 2024 - building up a new community here!
He will conduct his doctoral research under my supervision on the project “Understanding the structure of interphase chromosomes.”
#statistical_physics #polymers #chromatin #MD_simulations

He will conduct his doctoral research under my supervision on the project “Understanding the structure of interphase chromosomes.”
#statistical_physics #polymers #chromatin #MD_simulations
Pioneers of HTM will share their insights!
#Biosensors #HTM
More info: sciforum.net/event/Sensor...

Pioneers of HTM will share their insights!
#Biosensors #HTM
More info: sciforum.net/event/Sensor...
Join us next Friday 17 Oct. for “Mechanics, Dynamics and Shape in Oocytes” by Dr. Bart Vos — a deep dive into how physical forces and cell mechanics shape early development.
Discover the emerging field of mechanobiology, where physics meets life.

Join us next Friday 17 Oct. for “Mechanics, Dynamics and Shape in Oocytes” by Dr. Bart Vos — a deep dive into how physical forces and cell mechanics shape early development.
Discover the emerging field of mechanobiology, where physics meets life.

arxiv.org/abs/2510.07949

arxiv.org/abs/2510.07949

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
@cienciasuam.bsky.social @uam.es
Enrico Carlon @ecarlon.bsky.social from KU Leuven
will talk about DNA mechanics beyond the twistable wormlike chain. See you there!
ifimac.uam.es/ifimac-semin...

@cienciasuam.bsky.social @uam.es
Enrico Carlon @ecarlon.bsky.social from KU Leuven
will talk about DNA mechanics beyond the twistable wormlike chain. See you there!
ifimac.uam.es/ifimac-semin...
Open position for PhD Student Studying Vitro Single-molecule Transport Across Peroxisome Mimics
careers.tudelft.nl/job/Delft-Ph...

Open position for PhD Student Studying Vitro Single-molecule Transport Across Peroxisome Mimics
careers.tudelft.nl/job/Delft-Ph...
arxiv.org/abs/2508.15091

arxiv.org/abs/2508.15091
Great program and organization!
userswww.pd.infn.it/~orlandin/Ve...

Great program and organization!
userswww.pd.infn.it/~orlandin/Ve...
arxiv.org/abs/2507.00520

arxiv.org/abs/2507.00520
workingat.vu.nl/vacancies/po...
workingat.vu.nl/vacancies/po...
Biophysical principles of chromosome organisation
Info and registration:
fys.kuleuven.be/english/coll...

Biophysical principles of chromosome organisation
Info and registration:
fys.kuleuven.be/english/coll...

More information: www.biozentrum.unibas.ch/education/de...
#Master #Students #Research #Physics #Mathematics #ComputerScience #Engineering #Basel #Switzerland

More information: www.biozentrum.unibas.ch/education/de...
#Master #Students #Research #Physics #Mathematics #ComputerScience #Engineering #Basel #Switzerland


We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
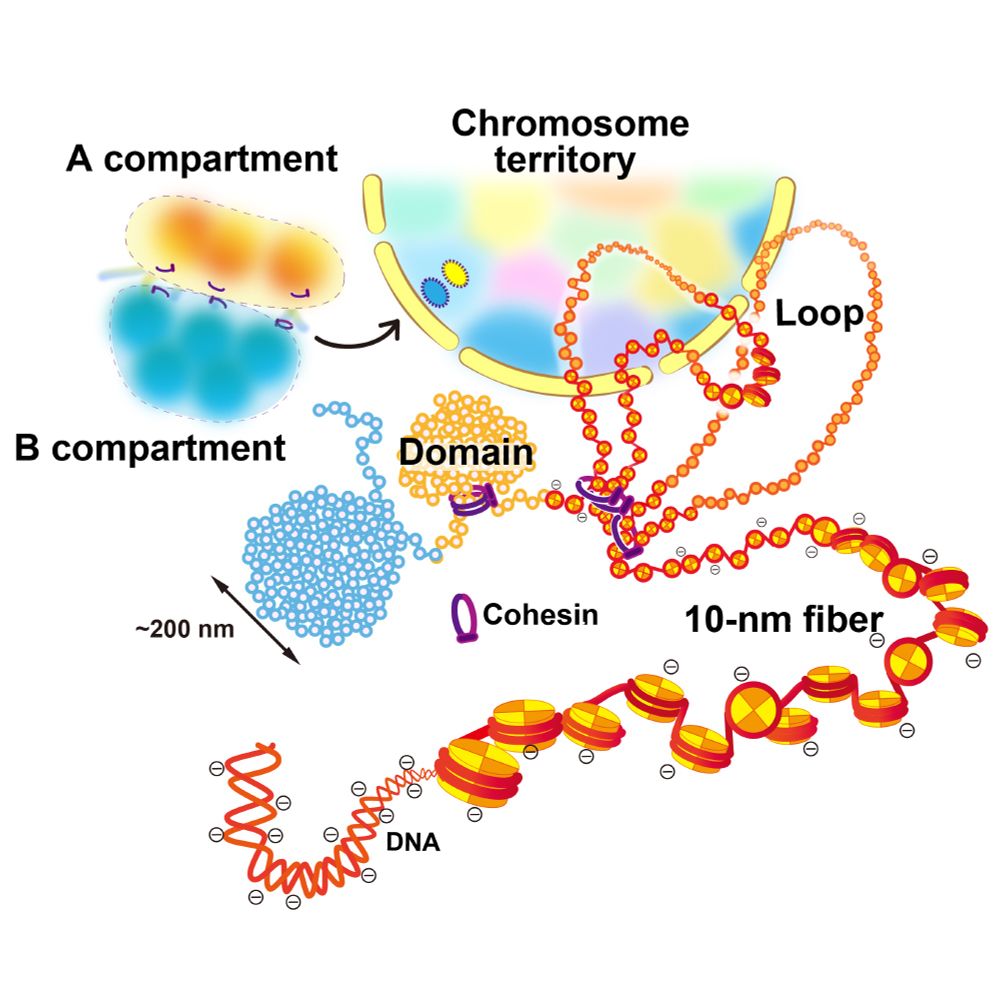
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
doi.org/10.1063/5.02...
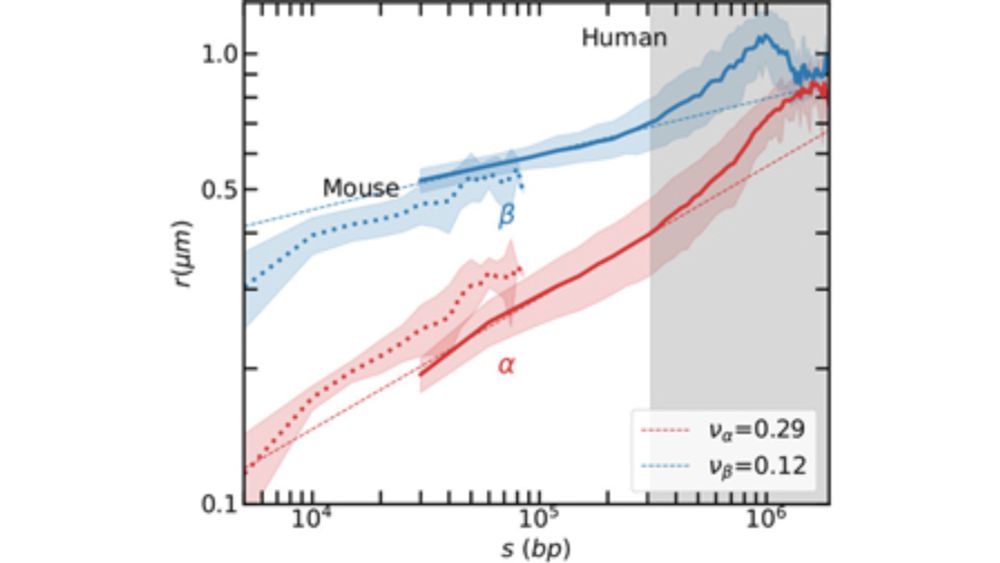
doi.org/10.1063/5.02...
Check our latest work with Mike Rosen and Sy Redding. We explore how changes in linker DNA length (as small as 1 bp) fine-tune chromatin structure, between order and disorder, and the properties of chromatin droplets



Check our latest work with Mike Rosen and Sy Redding. We explore how changes in linker DNA length (as small as 1 bp) fine-tune chromatin structure, between order and disorder, and the properties of chromatin droplets

