
Data with pasta. arxiv.org/pdf/2412.01561
Data with pasta. arxiv.org/pdf/2412.01561
www.cell.com/cell/fulltex...
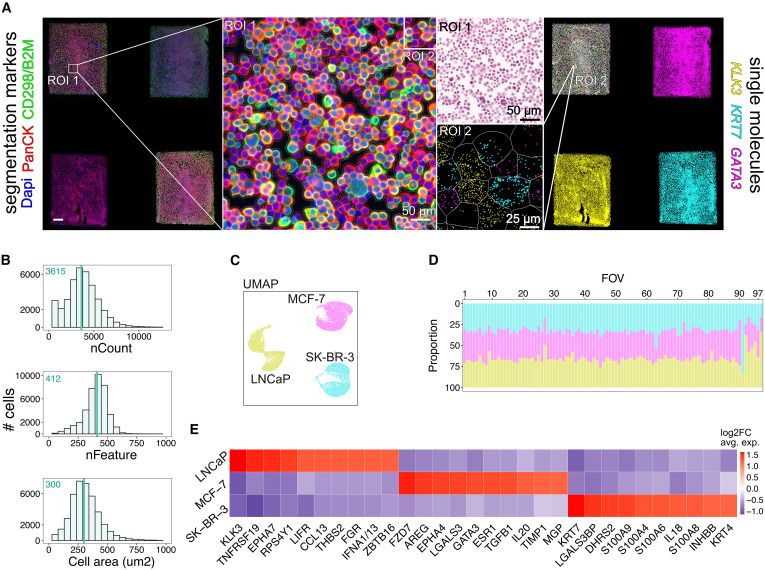
www.cell.com/cell/fulltex...
www.nature.com/articles/s41...
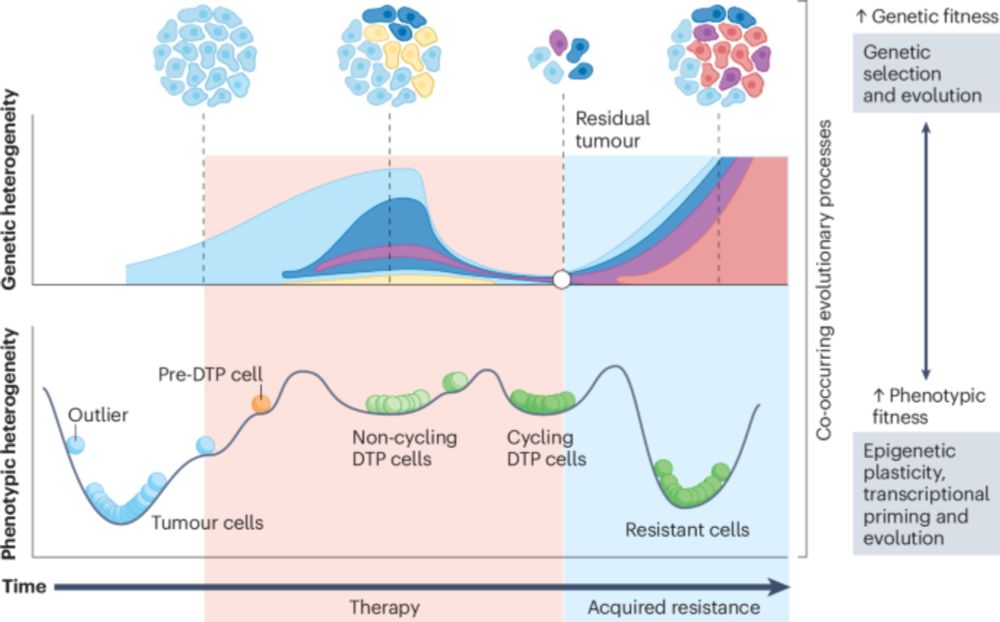
www.nature.com/articles/s41...
Exciting postdoc joint with @hartjackson.bsky.social developing machine learning methods for the next generation of spatial histopathology technologies joint with industry partners in the Bay Area.
👉 apply.interfolio.com/168307
Exciting postdoc joint with @hartjackson.bsky.social developing machine learning methods for the next generation of spatial histopathology technologies joint with industry partners in the Bay Area.
👉 apply.interfolio.com/168307
@davisjmcc.bsky.social
www.biorxiv.org/content/10.1...
@davisjmcc.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Are you a new investigator (<5 years) with a research group? Think your work would make an amazing logo? We’re giving away a FREE branding package (valued up to $1,000 CAD) to one lucky research group!
docs.google.com/forms/d/e/1F...

Are you a new investigator (<5 years) with a research group? Think your work would make an amazing logo? We’re giving away a FREE branding package (valued up to $1,000 CAD) to one lucky research group!
docs.google.com/forms/d/e/1F...
www.nature.com/articles/s41...
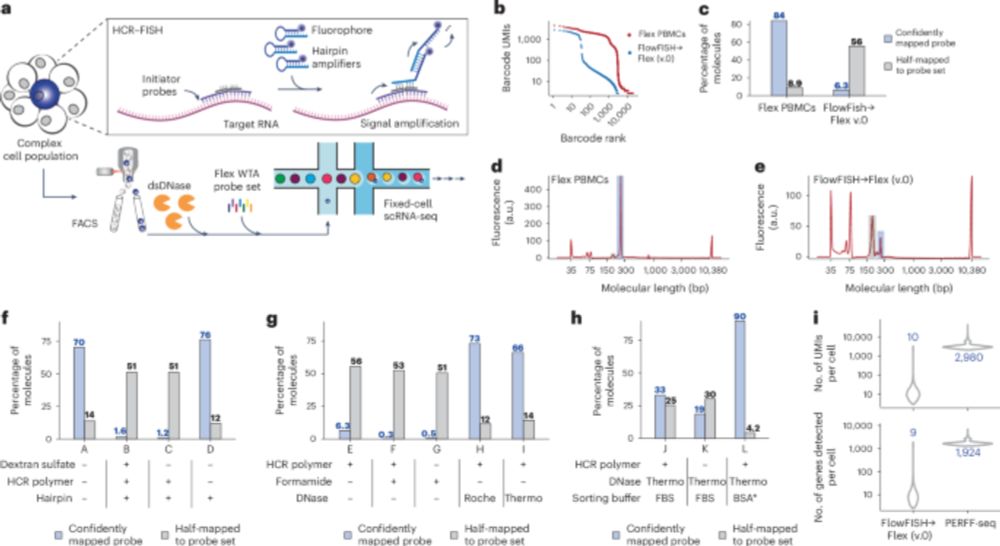
www.nature.com/articles/s41...
1/
www.nature.com/articles/s41...
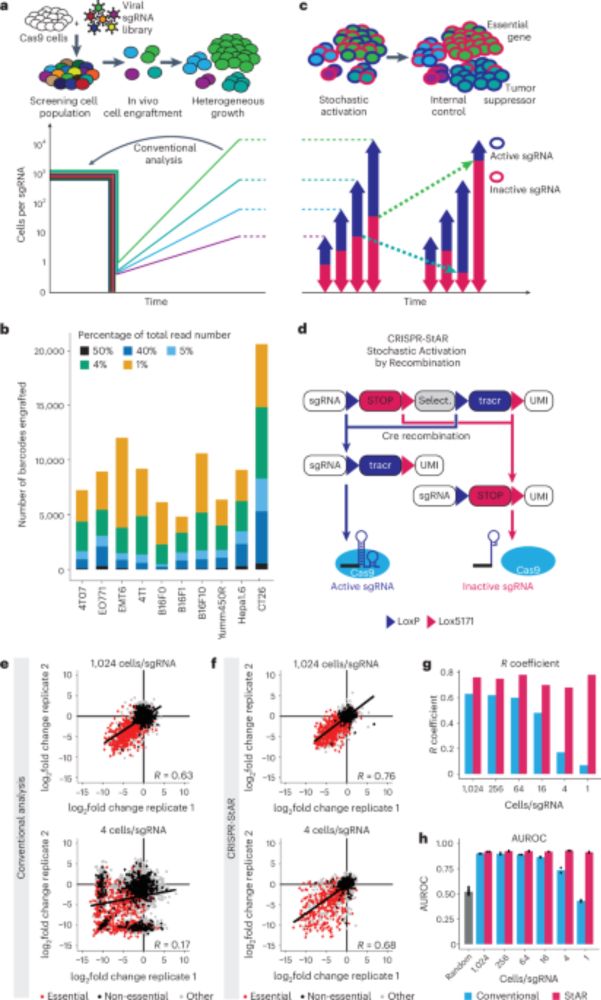
1/
www.nature.com/articles/s41...
This was an incredible team effort w/ @jennifergarrison.bsky.social #TammyLan @davidsebfischer.bsky.social #AlisonKochersberger #RuthRaichur #SophiaSzady
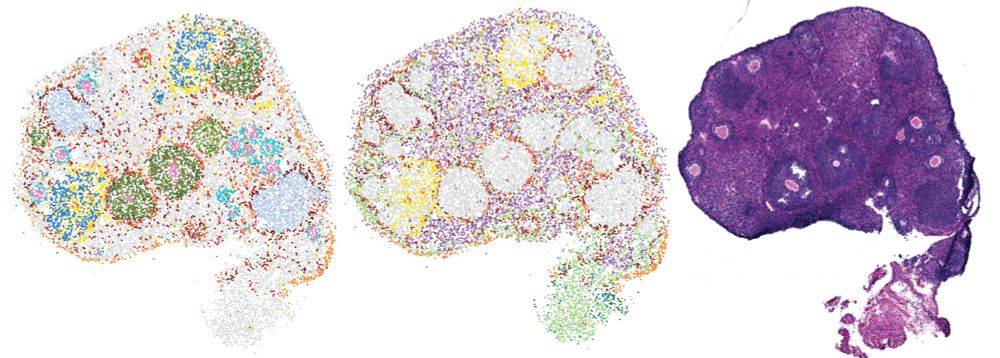
This was an incredible team effort w/ @jennifergarrison.bsky.social #TammyLan @davidsebfischer.bsky.social #AlisonKochersberger #RuthRaichur #SophiaSzady

Do you ever wonder how the #TME in #HGSC changes during chemotherapy? Check it here: www.sciencedirect.com/science/arti... and let’s dive into the highlights together #spatialbiology #ovariancancer #cancerresarch
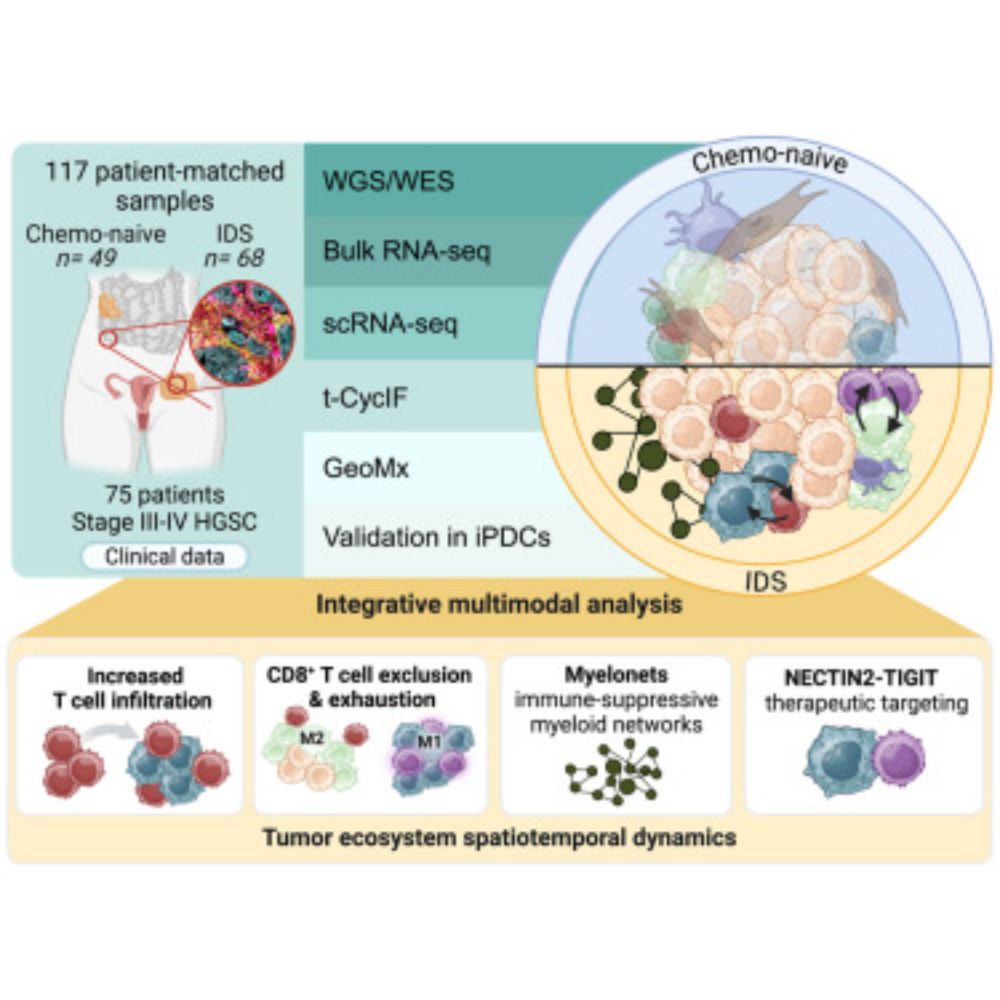
Do you ever wonder how the #TME in #HGSC changes during chemotherapy? Check it here: www.sciencedirect.com/science/arti... and let’s dive into the highlights together #spatialbiology #ovariancancer #cancerresarch
The preprint is on biorxiv now, but you can find a lot more details at www.muspan.co.uk, including documentation, worked tutorials (an ever-expanding list), install guides, and how to get the code!
The preprint is on biorxiv now, but you can find a lot more details at www.muspan.co.uk, including documentation, worked tutorials (an ever-expanding list), install guides, and how to get the code!
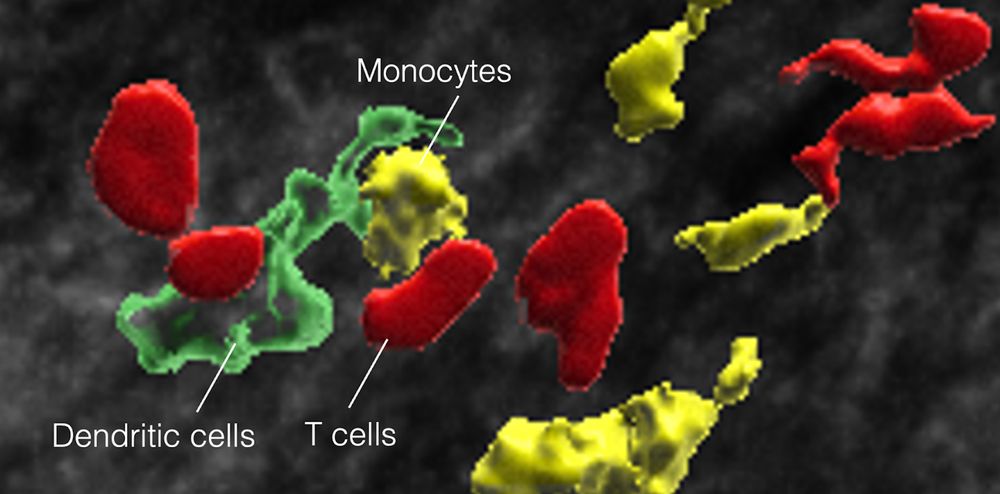
~300 researchers, oncologists, students, postdocs, patient advocates, and organizations supporting cancer research 🧪🔬⚕️
I hope you'll find these useful to build our community here
Pack 1: go.bsky.app/FEoaJMc
Pack 2: go.bsky.app/6EcjWNA

~300 researchers, oncologists, students, postdocs, patient advocates, and organizations supporting cancer research 🧪🔬⚕️
I hope you'll find these useful to build our community here
Pack 1: go.bsky.app/FEoaJMc
Pack 2: go.bsky.app/6EcjWNA
www.nature.com/articles/s41...

www.nature.com/articles/s41...
biorxiv.org/content/10.1101/2024.11.06.622266v1

biorxiv.org/content/10.1101/2024.11.06.622266v1



