







🔗 doi.org/10.1093/gbe/evaf242
📸 Evan D’Alessandro
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf242
📸 Evan D’Alessandro
#genome #evolution

At the PRBB Sustainability Seminar, Javier del Campo warns about the misuse of science to gain prestige or profit and highlights the social power of the scientific community.
More: tuit.cat/uvQp2

At the PRBB Sustainability Seminar, Javier del Campo warns about the misuse of science to gain prestige or profit and highlights the social power of the scientific community.
More: tuit.cat/uvQp2
Thanks, Nikita Kulikov, Kim Joffroy, @ambonacolta.bsky.social @delcampolab.bsky.social for your hard work

Thanks, Nikita Kulikov, Kim Joffroy, @ambonacolta.bsky.social @delcampolab.bsky.social for your hard work
Join our online course, with Anna Luigi and @delcampolab.bsky.social in March
@qiime2.org #Rstats
www.physalia-courses.org/courses-work...

Join our online course, with Anna Luigi and @delcampolab.bsky.social in March
@qiime2.org #Rstats
www.physalia-courses.org/courses-work...




Your only answer as a PI: Sure 😬
The outcome? Sleep-deprived students and an excellent work on the #coral holobiont rhythmicity 🪸🦠⏰
#symbiosis #microbiome

Your only answer as a PI: Sure 😬
The outcome? Sleep-deprived students and an excellent work on the #coral holobiont rhythmicity 🪸🦠⏰
#symbiosis #microbiome






Huge thanks to this amazing team! @fonamental.bsky.social @polcapdevila.bsky.social @elpiratavell.bsky.social @medrecover.bsky.social over.bsky.social @icmcsic.bsky.social @szndohrn.bsky.social @ub.edu @ibe-barcelona.bsky.social
#MedCalRes @ageinves.bsky.social @cienciagob.bsky.social
Bryozoans are key habitat-formers, yet often overlooked in climate research.
Our work shows they’re at risk under future ocean conditions.
🌊 We need more long-term, multi-stressor studies in natural settings. Stay tuned for the ongoing HOLOCHANGE and MedAcidWarm projects!


Bryozoans are key habitat-formers, yet often overlooked in climate research.
Our work shows they’re at risk under future ocean conditions.
🌊 We need more long-term, multi-stressor studies in natural settings. Stay tuned for the ongoing HOLOCHANGE and MedAcidWarm projects!
This is the first study to document long-term microbiome and skeletal responses to combined ocean acidification & warming in bryozoans in the wild.
We reveal:
✅ Complex, species-specific strategies
⚠️ Signs of vulnerability under future ocean conditions
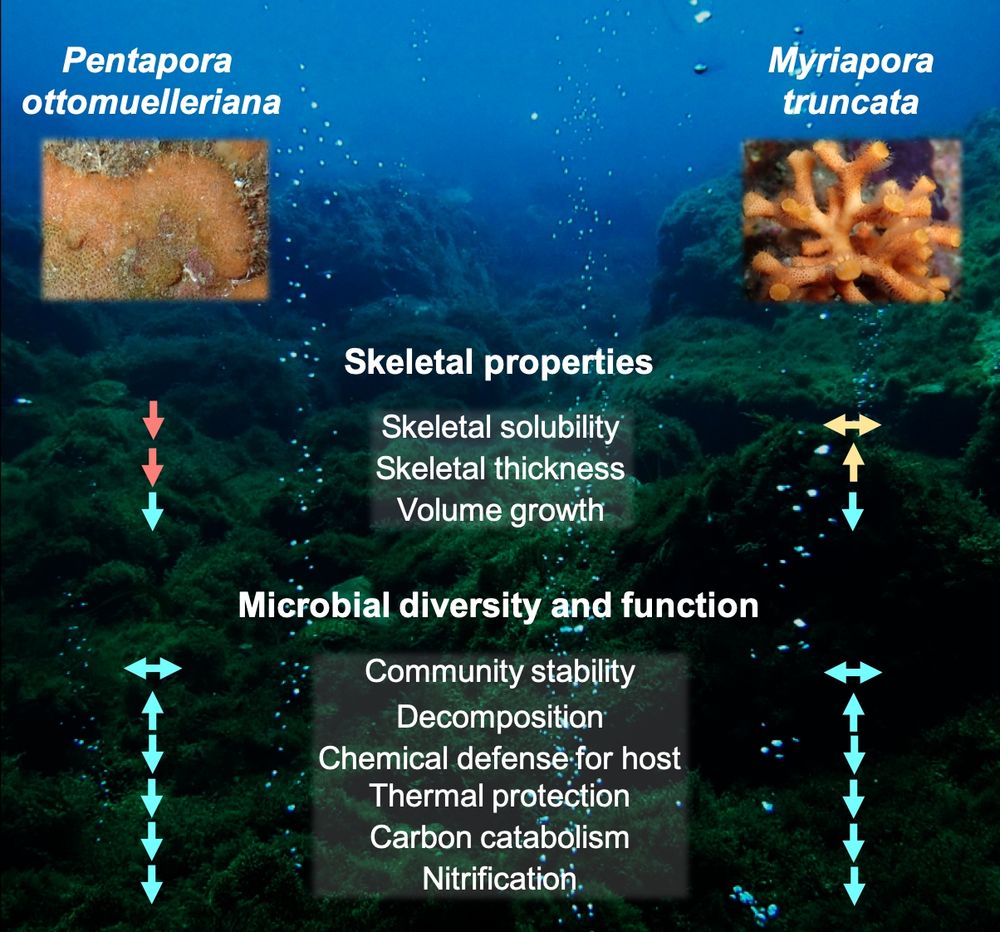
This is the first study to document long-term microbiome and skeletal responses to combined ocean acidification & warming in bryozoans in the wild.
We reveal:
✅ Complex, species-specific strategies
⚠️ Signs of vulnerability under future ocean conditions
What about long-term effects?
We monitored Pentapora populations over 5 years:
📉 Bryozoan cover declined
☠️ Necrosis increased
🌡️ Warming & acidification together accelerated mortality
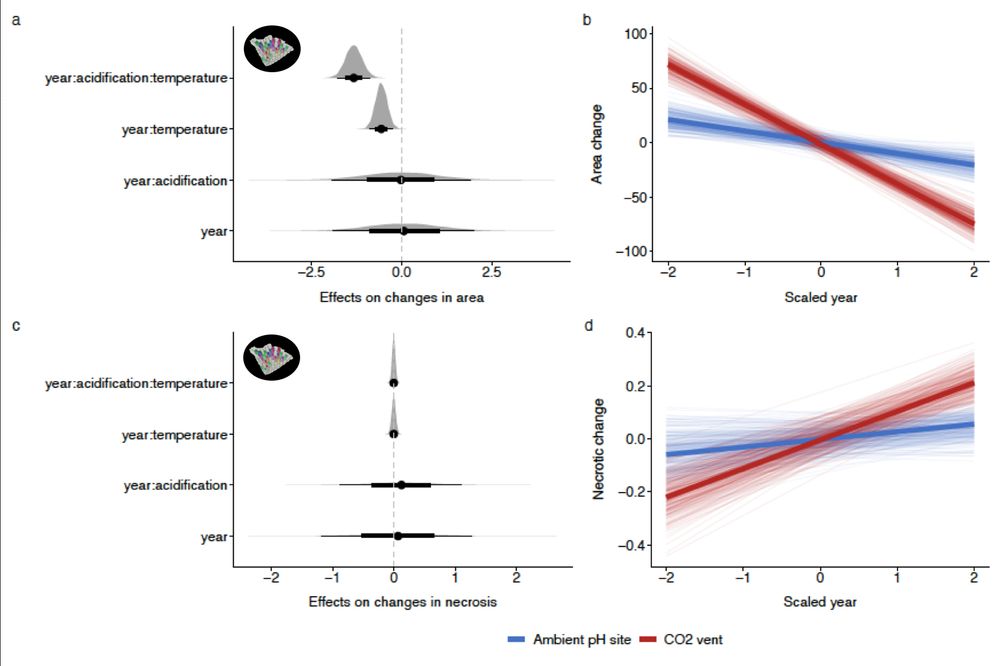
What about long-term effects?
We monitored Pentapora populations over 5 years:
📉 Bryozoan cover declined
☠️ Necrosis increased
🌡️ Warming & acidification together accelerated mortality
But not all are good news...
We also observed:
📉 Loss of key microbial genera
☠️Rise of opportunists & anaerobes
These microbial shifts may signal early dysbiosis — an early warning of declining host health.
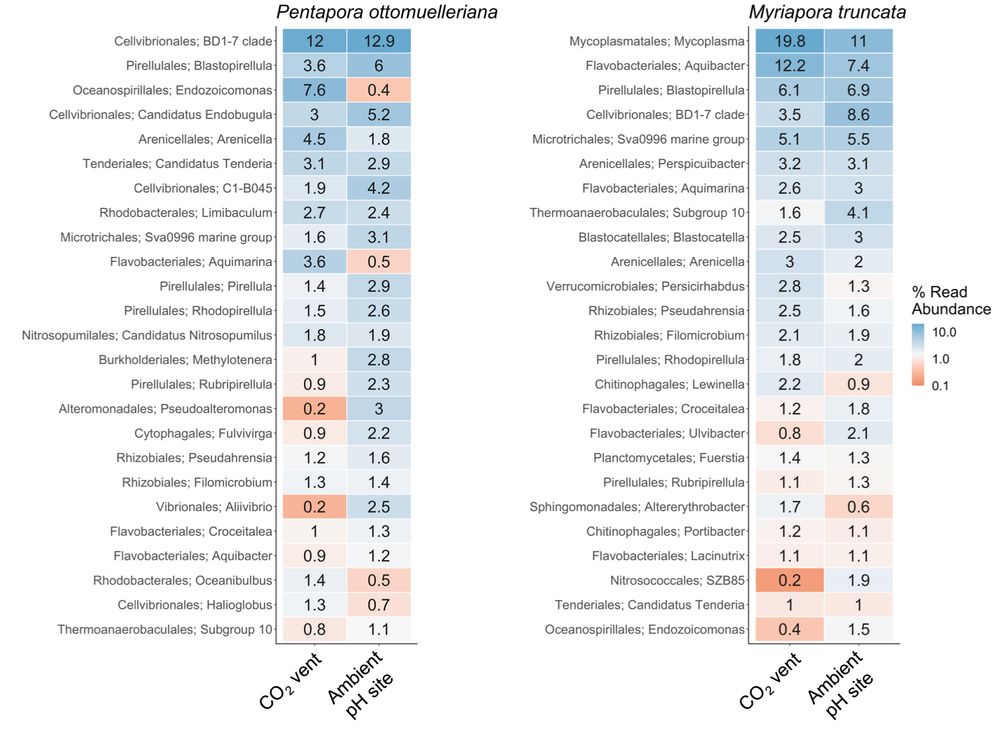
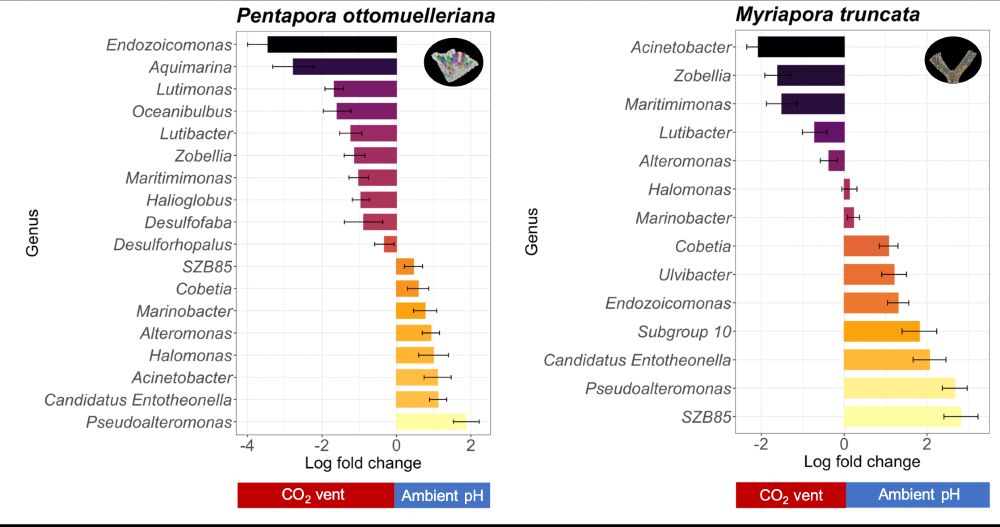
But not all are good news...
We also observed:
📉 Loss of key microbial genera
☠️Rise of opportunists & anaerobes
These microbial shifts may signal early dysbiosis — an early warning of declining host health.
🦠 Microbial communities:
We found species-specific core microbiomes, conserved across both pH conditions.
This stability suggests potential acclimatization capacity in both species despite environmental stress.
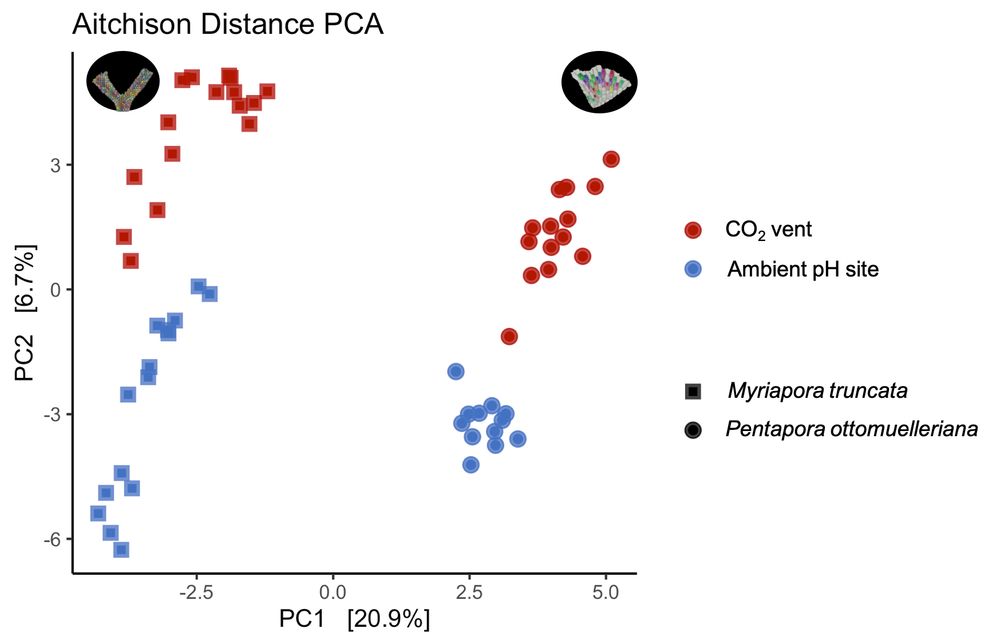
🦠 Microbial communities:
We found species-specific core microbiomes, conserved across both pH conditions.
This stability suggests potential acclimatization capacity in both species despite environmental stress.

