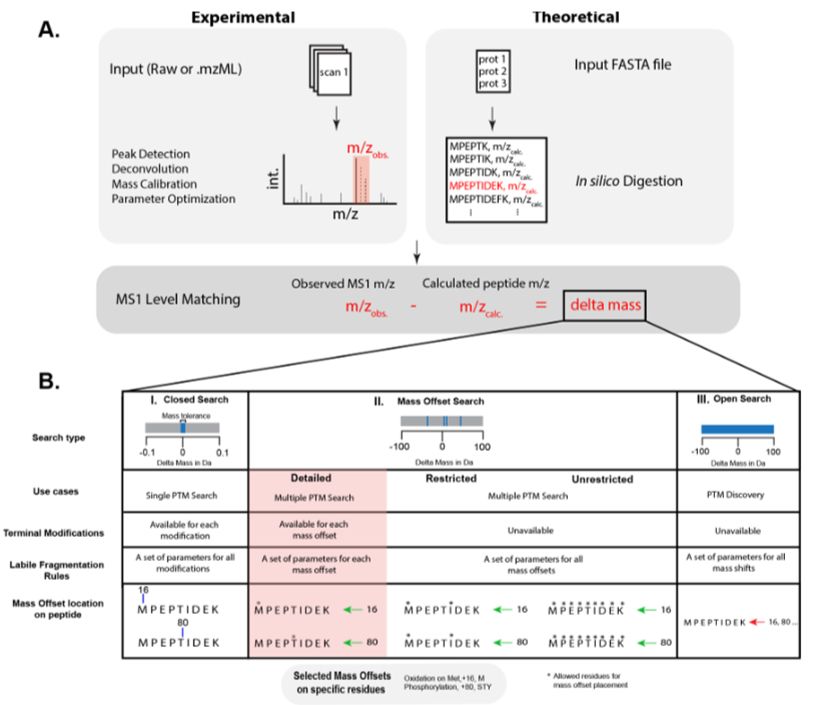
---
#proteomics #prot-paper
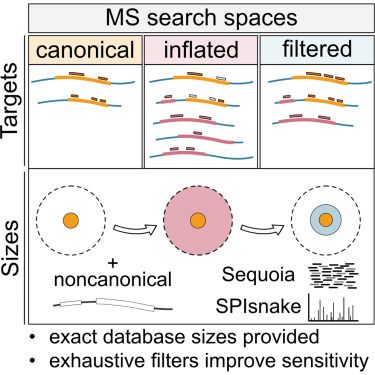
---
#proteomics #prot-paper
---
#proteomics #prot-paper
---
#proteomics #prot-paper
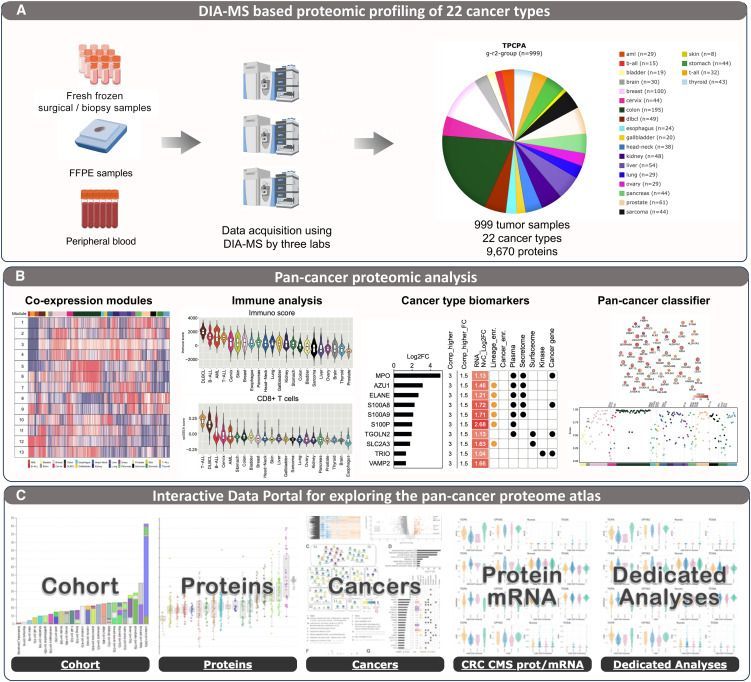
Science was excellent and the personal discussions enriching. I was charmed to reconnect with the French proteomics community, and the breeze and beauty of Saint Malo made it unforgettable.
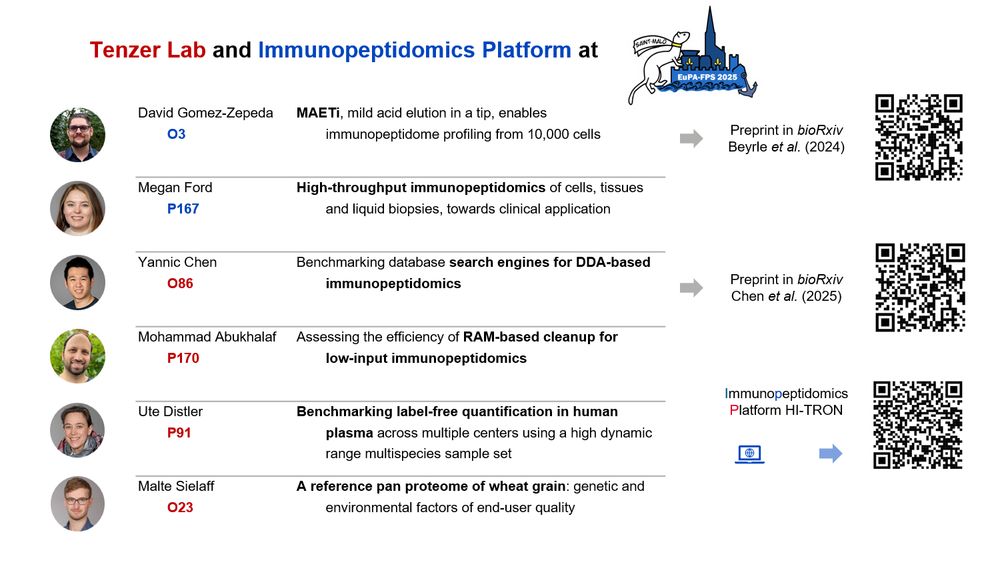
Science was excellent and the personal discussions enriching. I was charmed to reconnect with the French proteomics community, and the breeze and beauty of Saint Malo made it unforgettable.
---
#proteomics #prot-paper
---
#proteomics #prot-paper

---
#proteomics #prot-other

---
#proteomics #prot-other
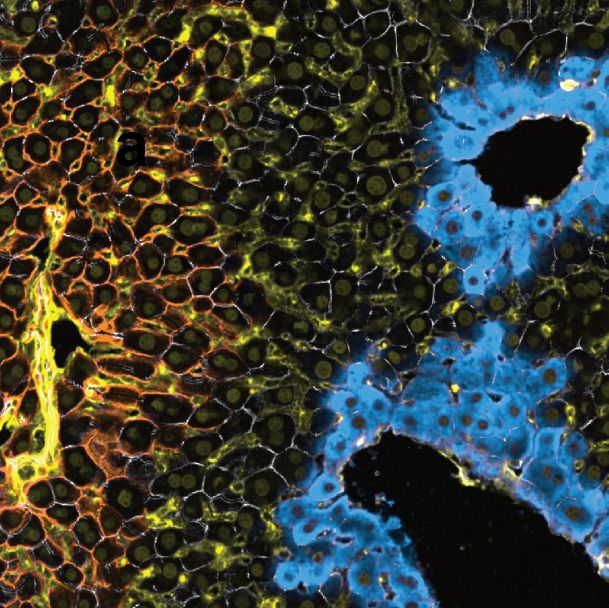
Let us take a tour through proteotoxic stress in intact human tissue — one hepatocyte at a time.
www.nature.com/articles/s41...
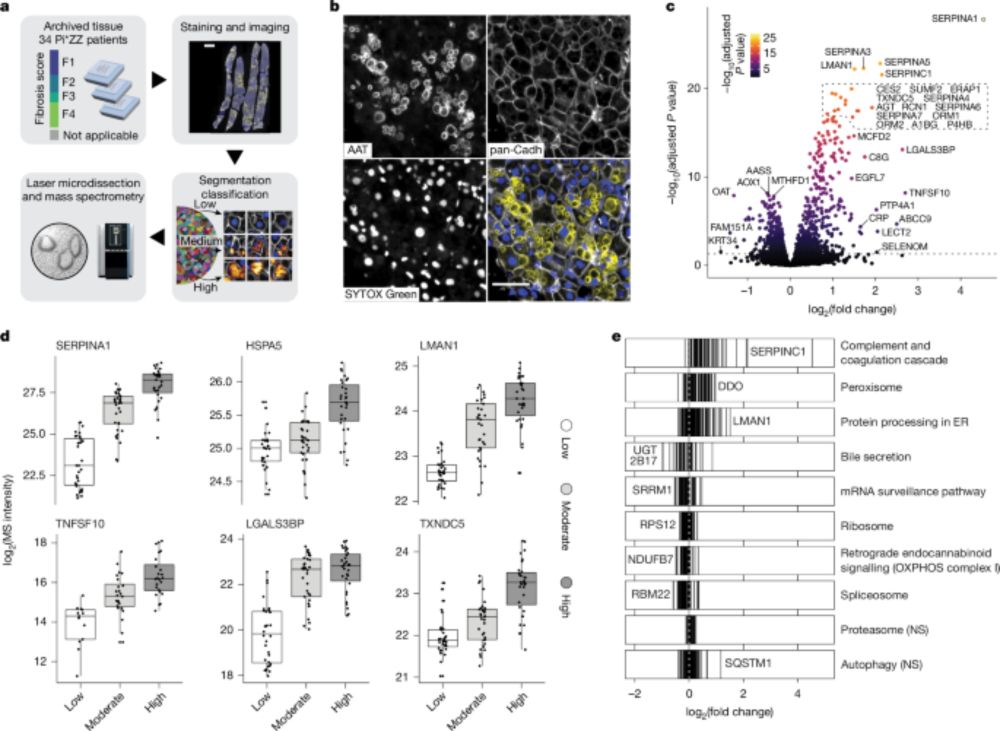
Let us take a tour through proteotoxic stress in intact human tissue — one hepatocyte at a time.
www.nature.com/articles/s41...
---
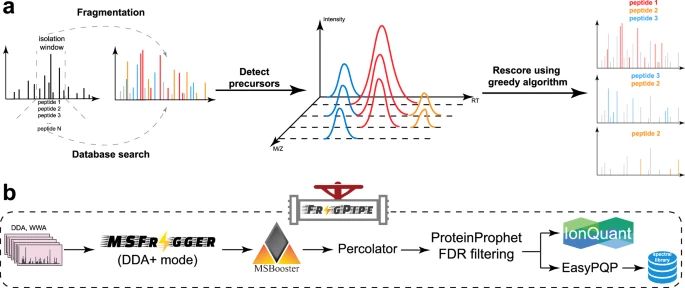
#proteomics #prot-paper
---
#proteomics #prot-paper
Join our webinar exploring the topic on April 24 here: www.evosep.com/webinars/web...
#teammassspec


pubs.acs.org/doi/10.1021/...
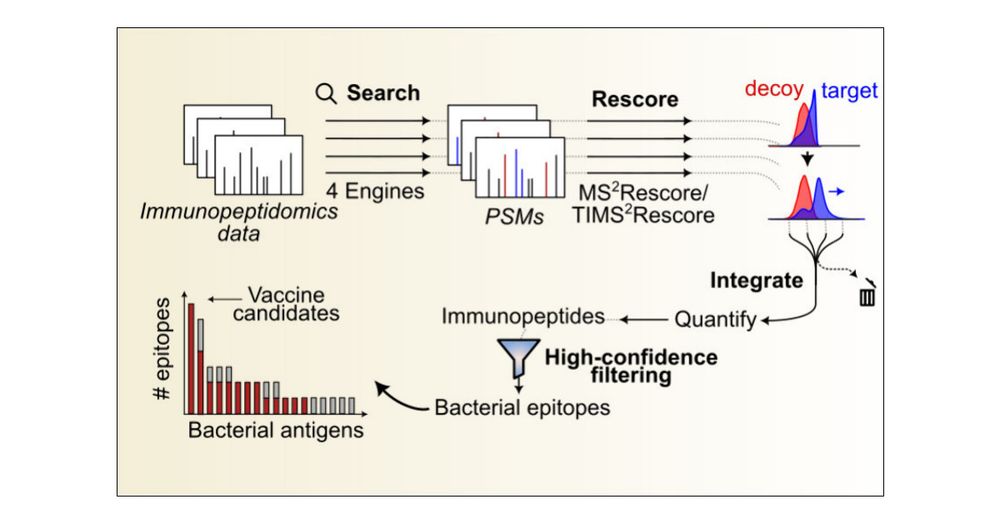
pubs.acs.org/doi/10.1021/...
www.nature.com/articles/s41...
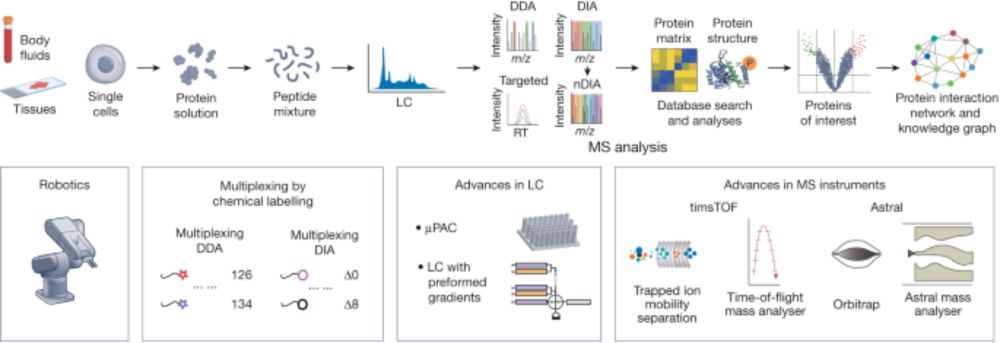
www.nature.com/articles/s41...
---
#proteomics #prot-preprint
---
#proteomics #prot-preprint
If you’re an expert in computational mass spectrometry, machine-learning-driven omics & high-throughput molecule identification, apply now for our tenure-track position at @univie.ac.at
jobs.univie.ac.at/job/Tenure-T...
#Hiring #MassSpec #Bioinformatics #Jobs
If you’re an expert in computational mass spectrometry, machine-learning-driven omics & high-throughput molecule identification, apply now for our tenure-track position at @univie.ac.at
jobs.univie.ac.at/job/Tenure-T...
#Hiring #MassSpec #Bioinformatics #Jobs
We are happy to support his application at the Immunopeptidomics Platform HI-TRON and hope the fund will make this breakthrough project possible in collab with David from @lukasbunse.bsky.social lab at @dkfz.bsky.social
Cantón!
David is a biotechnologist by training who specialized in cancer research during his master’s studies and is now pursuing his PhD at DKFZ-German Cancer Research Center.

We are happy to support his application at the Immunopeptidomics Platform HI-TRON and hope the fund will make this breakthrough project possible in collab with David from @lukasbunse.bsky.social lab at @dkfz.bsky.social
---
#proteomics #prot-other

---
#proteomics #prot-other

Just like the past years, we, led by Dr. Christina Ludwig, are part of the organization of the amazing Brixen Proteomics Summer School.
The first announcement is going on since a couple of days and many more exciting news will follow again soon.

Just like the past years, we, led by Dr. Christina Ludwig, are part of the organization of the amazing Brixen Proteomics Summer School.
The first announcement is going on since a couple of days and many more exciting news will follow again soon.

