Many thanks to the committee members Nick Irwin, Nacho Maeso and Sara Sdelci @sdelcilab.bsky.social!

Many thanks to the committee members Nick Irwin, Nacho Maeso and Sara Sdelci @sdelcilab.bsky.social!
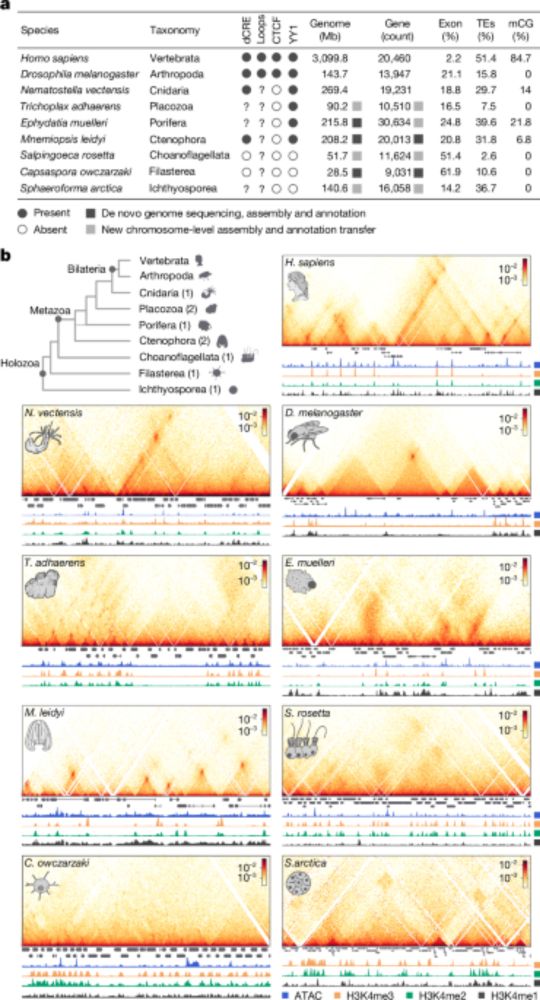
We developed iChIP2 to profile hPTMs across eukaryotes. Thanks to David and Julen, and specially to @seanamontgomery.bsky.social, the best teammate I could wish:)
Check out what we discovered about chromatin evolution!
We developed iChIP2 to profile hPTMs across eukaryotes. Thanks to David and Julen, and specially to @seanamontgomery.bsky.social, the best teammate I could wish:)
Check out what we discovered about chromatin evolution!

