Statistical Genetics @INRAE Toulouse
https://gestat.netlify.app/
Université Fédérale de Toulouse Midi-Pyrénées; École Nationale Vétérinaire de Toulouse; Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement; Génétique Physiologie et Systèmes d'Elevage • Genetic and phenotypic traits in livestock, Genetic Mapping and Diversity in Plants and Animals, Insect and Pesticide Research
Job description is here : urls.fr/FZNWJQ
Don't hesitate to contact me for more information !
Reposted by Bertrand Servin
Détails ici:
emploi.cnrs.fr/Offres/CDD/U...
Début 01/01/2026 pour 18 mois potentiellement renouvelable.
candidature jusqu'au 29/11
N’hésitez pas à diffuser largement
Reposted by Frédéric Delsuc, Bertrand Servin

These two males belong to different species—but share the same mother. How? Why?
To celebrate the print release of our last paper in this week’s @nature.com (issue 8084), here’s a thread summarizing the results. Why? Let’s dive in🧵👇 www.nature.com/articles/s41...
Reposted by Bertrand Servin
The next #PopGroup meeting will take place in Lille 🍟, France, 7–9 January 2026 – just 1 hour by train from London, Brussels, and Paris.
This year, PopGroup will also host ALPHY, the annual meeting of Evolutionary Genomics.
More info: populationgeneticsgroup.org.uk
See you there !
Reposted by Philip Nel, Bertrand Servin

), we find the opposite: animals exchange genes more, and for longer, than plants
Reposted by Jason Stajich, Bertrand Servin

Reposted by Bertrand Servin
After 100 years of #popgen people does not seem to be totally agree on what LD is?
en.wikipedia.org/wiki/Talk:Li...
More details here : jobs.inrae.fr/en/ot-25908 and even more after contacting us :) Applications are open !
Reposted by Bertrand Servin
Out now in Cell: Our new study uncovers the ancient origins of a genetic mutation that protects against HIV — and rewrites the story of its surprising high frequency in Europe.
Link: doi.org/10.1016/j.ce...
Let’s dig in (popular science first, jump to 16 for technical details)
Reposted by Bertrand Servin


Serving as Editor-in-Chief at Science was fascinating. I greatly enjoyed working with talented and committed editorial, news, graphics, and production staff. But the inside look into scientific publishing and AAAS was also deeply disillusioning.
Reposted by Bertrand Servin
ecoevorxiv.org/repository/v...
evolbiol.peercommunityin.org/articles/rec...
#phenplas #quantgen
Reposted by Bertrand Servin

genomestake.substack.com/p/genome-edi...
Reposted by Bertrand Servin
Reposted by Bertrand Servin

This was a ton of work.
medrxiv.org/cgi/content/...
1/
Reposted by Paul Ekblom, Bertrand Servin

have you written your Senator to let them know?
Reposted by Bertrand Servin, Victor A. Albert

www.nature.com/articles/s41...
Reposted by Bertrand Servin
Reposted by Bertrand Servin
Reposted by Bertrand Servin
All quotes are from "Good Thinking", store.doverpublications.com/products/978....
Reposted by Bertrand Servin

Reposted by Bertrand Servin, Greta Bocedi

#postdoc #genomics #insects #inversions
genomicsofoutbreaks.com/?page_id=141
Reposted by Bertrand Servin

Reposted by Bertrand Servin
Reposted by Greg Barton, Bertrand Servin
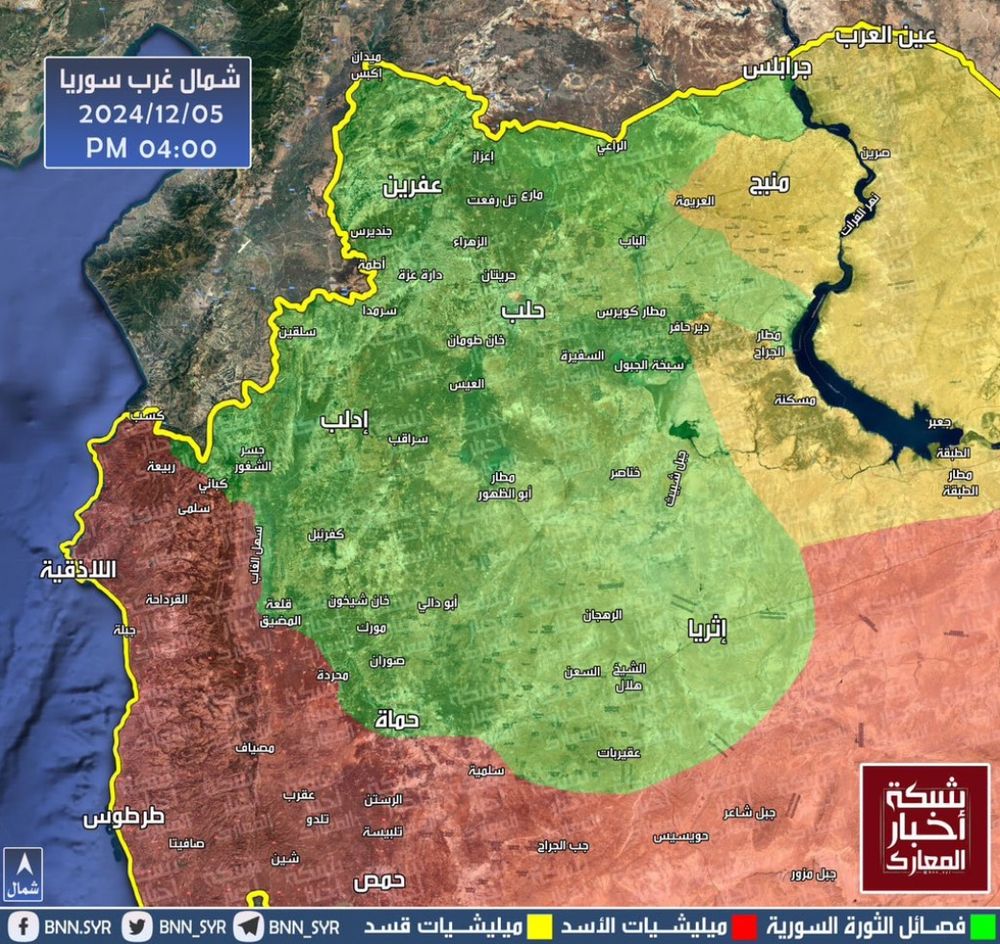
Reposted by Bertrand Servin
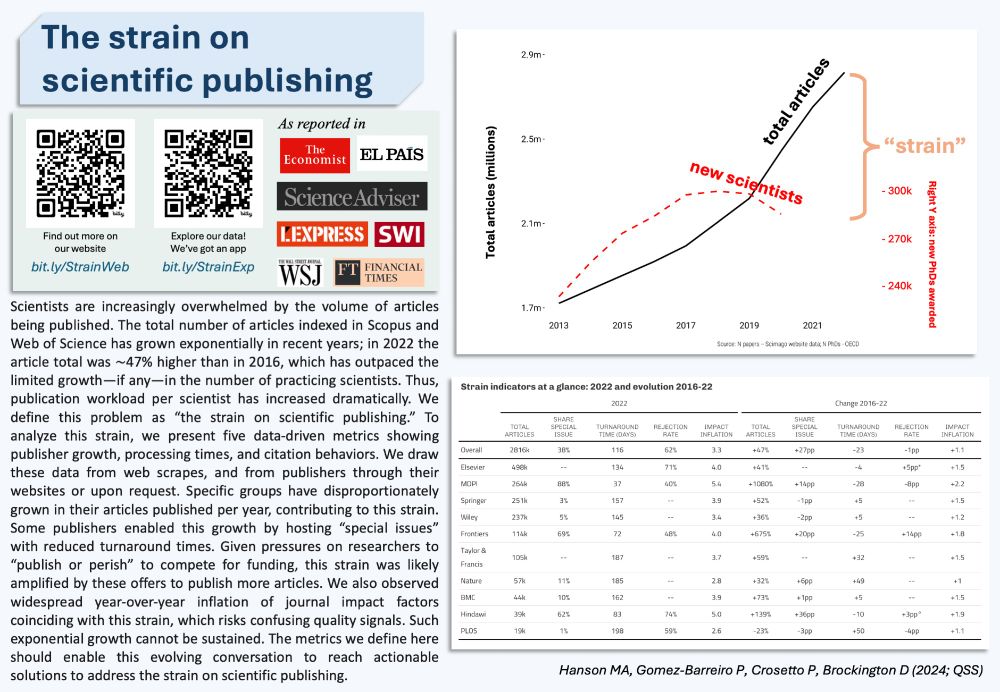
direct.mit.edu/qss/article/...
A 🧵 1/n
#AcademicSky #PhDchat #ScientificPublishing #SciPub
Reposted by Bertrand Servin

1/7
openreview.net/forum?id=H7m...
Reposted by Bertrand Servin