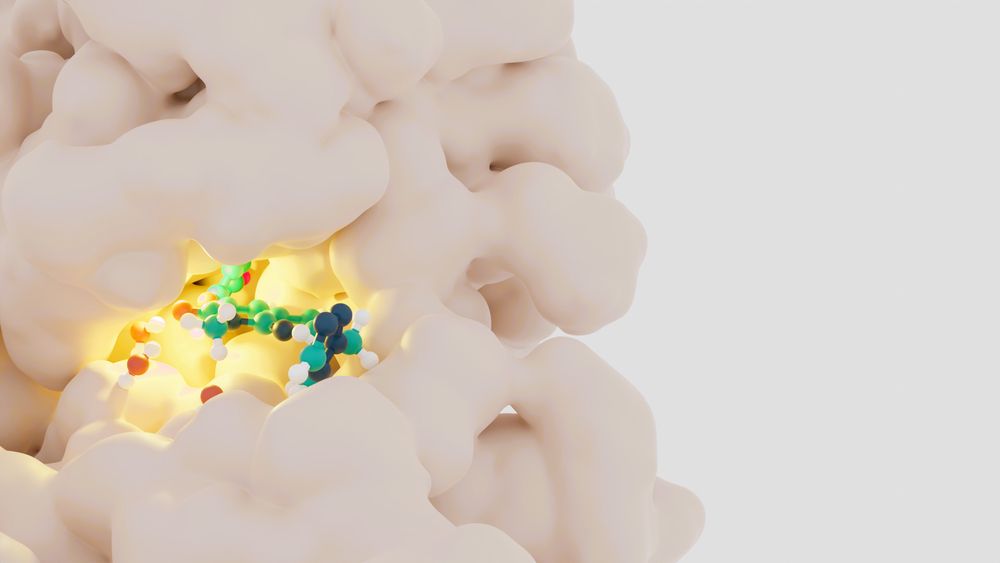
Docking remains essential in early-stage drug discovery, but recent deep learning–based approaches still face limitations in generating...
Thread - (1/n)
At the #mdcBerlin, @al2na.bsky.social’s team built Flexynesis, a deep learning toolkit to guide precision cancer care.
Learn more:
👉 www.mdc-berlin.de/news/press/u... 👈

At the #mdcBerlin, @al2na.bsky.social’s team built Flexynesis, a deep learning toolkit to guide precision cancer care.
Learn more:
👉 www.mdc-berlin.de/news/press/u... 👈
Docking remains essential in early-stage drug discovery, but recent deep learning–based approaches still face limitations in generating...
Thread - (1/n)
Docking remains essential in early-stage drug discovery, but recent deep learning–based approaches still face limitations in generating...
Thread - (1/n)
Docking remains essential in early-stage drug discovery, but recent deep learning–based approaches still face limitations in generating...
Thread - (1/n)
📢Register for the #ISCO'25 Conference "Innovations in #SingleCell #OMICS" in Berlin!
🗓️ 12-13 May 2025
🎤 Fantastic Keynote and Invited Speakers
🫵🏿 Many slots for talks: submit your abstract
🔗http://isco-conference.eu
Please spread the word!

📢Register for the #ISCO'25 Conference "Innovations in #SingleCell #OMICS" in Berlin!
🗓️ 12-13 May 2025
🎤 Fantastic Keynote and Invited Speakers
🫵🏿 Many slots for talks: submit your abstract
🔗http://isco-conference.eu
Please spread the word!
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

