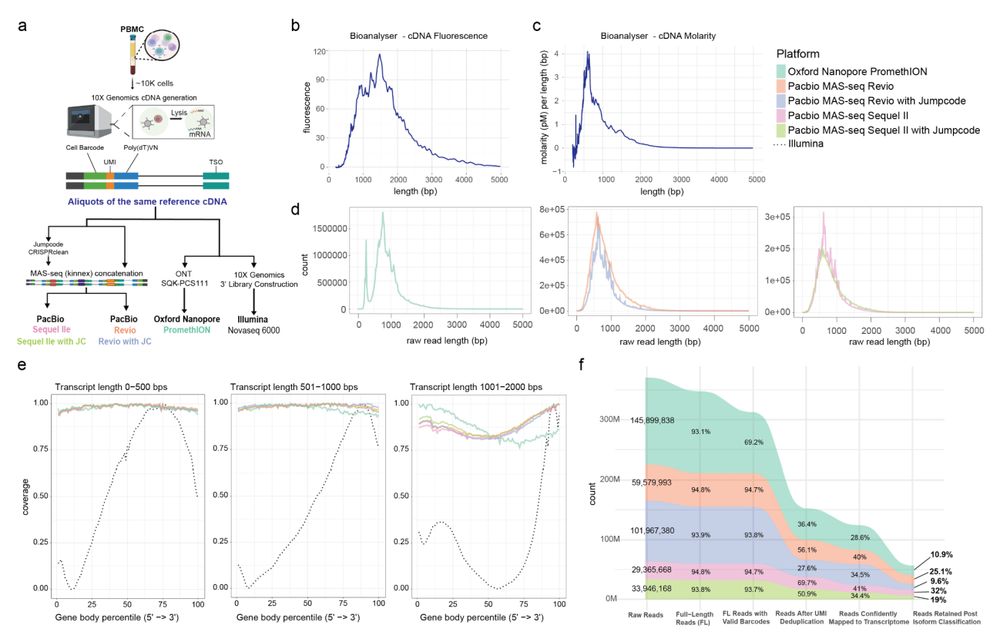
A comparison of long-read single-cell transcriptomic approaches
Long-read sequencing enables the incorporation of isoform-level expression into single-cell transcriptomic studies, offering detail beyond those accessible with short-read methods. Although insightful, these approaches have typically been costly and yielded limited data for each individual cell. Recent advances in library preparation approaches and sequencing throughput have brought long-read single-cell studies closer to the mainstream. Here, we present a comparative analysis of commercial approaches for single-cell long-read sequencing. We have performed parallel analyses of the same cDNA material, generated using the 10X genomics platform, on Illumina short-read, and PacBio and Oxford Nanopore long-read platforms. We also demonstrate the impact of CRISPR-based depletion of libraries, to remove highly expressed transcripts, prior to long-read sequencing in these experiments. By analysing single-source cDNA libraries in parallel, we enable a direct comparison of each platform, evaluating standard metrics alongside concordance in clustering and cell type identification. While each approach generates usable gene and isoform expression data, we identify limitations common across platforms, primarily linked to cDNA synthesis inefficiencies and read filtering strategies. Our work demonstrates the increasing utility of single-cell long-read sequencing for isoform-resolved analyses, such as direct immunoglobulin chain reconstruction without additional amplification, and the detection of alternative splicing patterns across immune cell subtypes in CD45, a key gene for immune cell activation and differentiation. Our benchmarking of current platform options provides a foundation for researchers looking to adopt single-cell long-read sequencing into their transcriptomic studies, providing a framework for its integration into diverse biological questions. ### Competing Interest Statement A.P.C. is an inventor on patents filed by Oxford University Innovations for single-cell technologies and is a co-founders of Entelo Bio. All other authors declare no conflict of interest. Cancer Research UK, https://ror.org/054225q67, A26815 Biotechnology and Biological Sciences Research Council, https://ror.org/00cwqg982, BB/CCG1720/1, BB/CCG2220/1, BBS/E/T/000PR9816, BBX011070/1, BB/V016156/1, BB/T008717/1 UK Research and Innovation, EP/X035913/1, 10098097 Biotechnology and Biological Sciences Research Council, BB/M011216/1, BBS/E/ER/23NB0006 Medical Research Council, MR/V010182/1