
Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

I'm so excited about this!
www.biorxiv.org/content/10.1...

I'm so excited about this!
www.biorxiv.org/content/10.1...
@davisjmcc.bsky.social
www.biorxiv.org/content/10.1...
@davisjmcc.bsky.social
www.biorxiv.org/content/10.1...
doi.org/10.1038/s415...
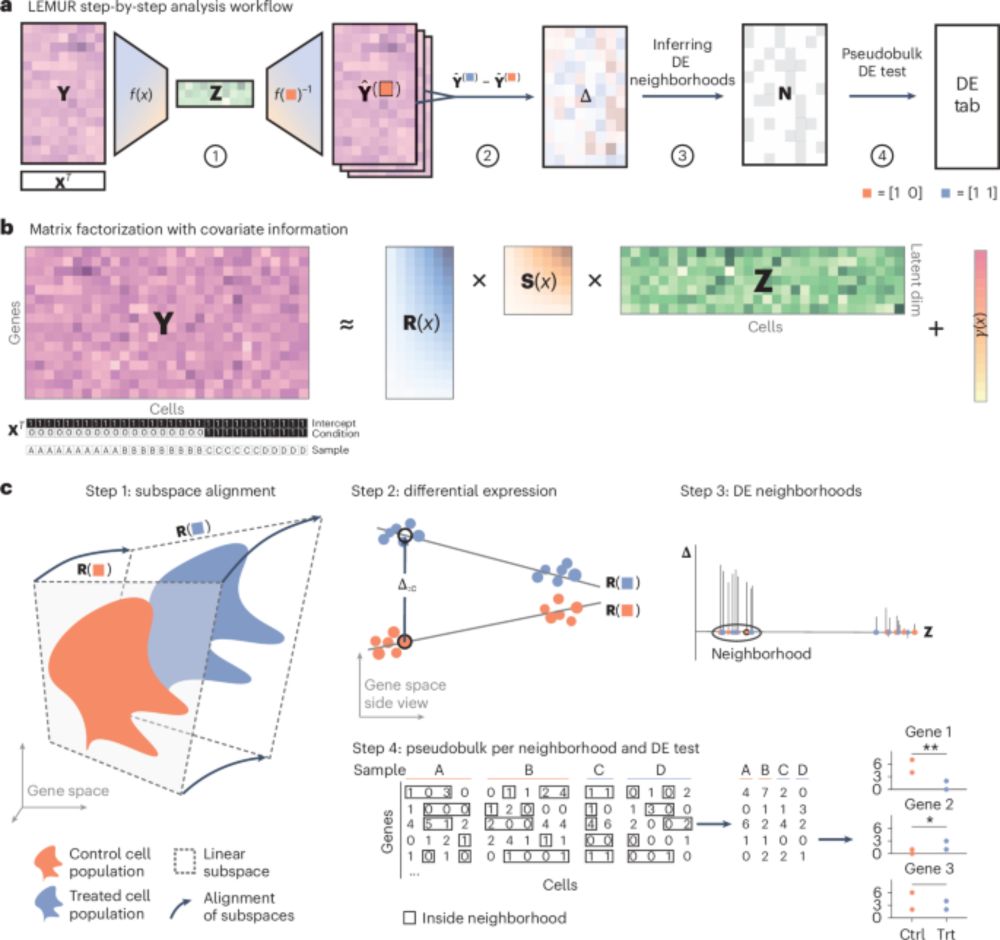
doi.org/10.1038/s415...
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻


Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻

Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻

