






ceryshafana.com

ceryshafana.com



10/n
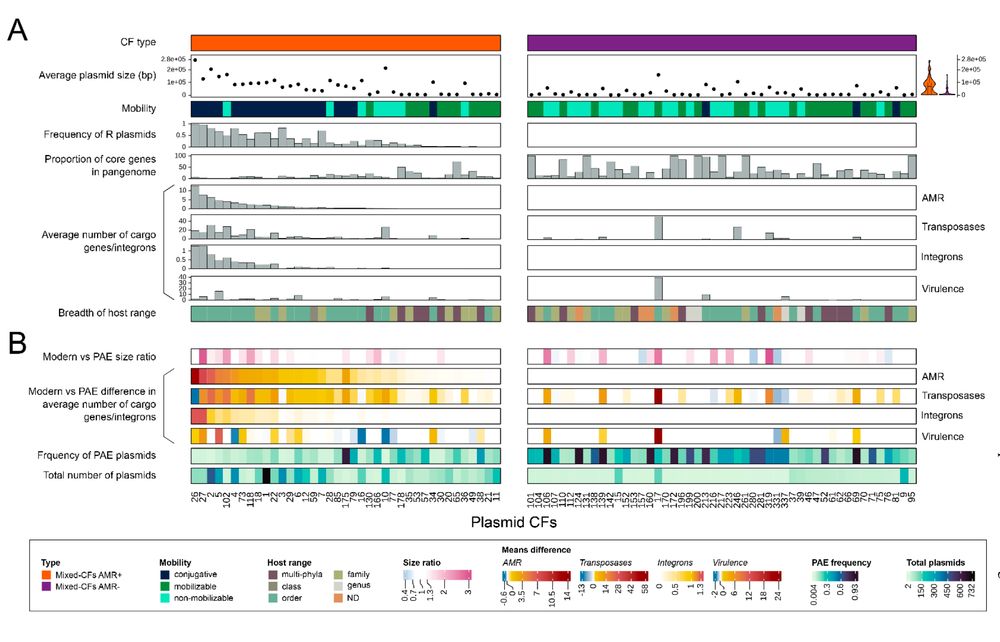
10/n
6/n

6/n
Thread 1/n

Thread 1/n
millions of genomes, allowing full alignment. Figure shows: time taken to align 2 genes, as size of the database grows to 1 million
genomes LexicMap (green) is faster (lower on y axis; log scale!!) and uses less RAM (smaller blobs). 4/n

millions of genomes, allowing full alignment. Figure shows: time taken to align 2 genes, as size of the database grows to 1 million
genomes LexicMap (green) is faster (lower on y axis; log scale!!) and uses less RAM (smaller blobs). 4/n
of mapping to a single reference over the years,
but algorithms for BLAST-ing against a huge database have not
been able to scale with data growth 2/n
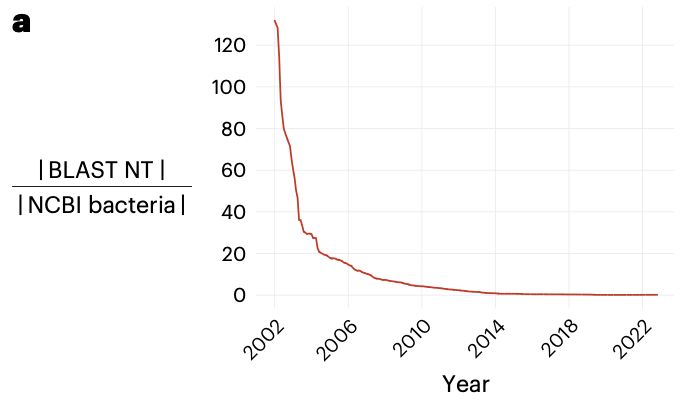
of mapping to a single reference over the years,
but algorithms for BLAST-ing against a huge database have not
been able to scale with data growth 2/n





#ABPHM25

#ABPHM25

and 99.6% for AMRFP Flye, 99.3% and 99.6% for AMRFP Raven, and 99.6% and 99.6% for AMRFP Unicycler.
15/n

and 99.6% for AMRFP Flye, 99.3% and 99.6% for AMRFP Raven, and 99.6% and 99.6% for AMRFP Unicycler.
15/n

11/n

11/n


