The tetrode follows the predicted path, and the gamma profiles match the silicon probe data beautifully at each step.
See one going from CA1 pyr to DG midmol below!

The tetrode follows the predicted path, and the gamma profiles match the silicon probe data beautifully at each step.
See one going from CA1 pyr to DG midmol below!
CA3 terminal stimulation strongly activated pyramidale (PyrInt) and radiatum (rad) interneurons, while EC terminal stimulation strongly drove LM interneurons.
(10/13)
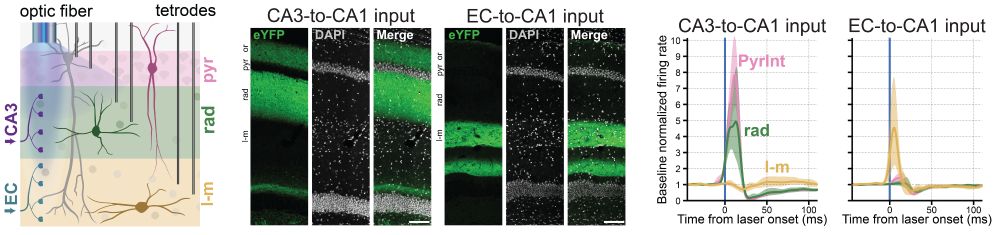
CA3 terminal stimulation strongly activated pyramidale (PyrInt) and radiatum (rad) interneurons, while EC terminal stimulation strongly drove LM interneurons.
(10/13)
Firing behavior was layer-specific; e.g., LM interneurons were barely active in ripples.
Preferred theta phase also varied across layers.
(9/13)

Firing behavior was layer-specific; e.g., LM interneurons were barely active in ripples.
Preferred theta phase also varied across layers.
(9/13)
By projecting tetrode LFPs from several mice onto the embedding (estimating each tetrode’s “depth”), we recover gradually changing theta and ripple waveforms, as if recorded with a single linear probe.
No need for alignment or histology. Just the signals and the embedding.
(8/13)

By projecting tetrode LFPs from several mice onto the embedding (estimating each tetrode’s “depth”), we recover gradually changing theta and ripple waveforms, as if recorded with a single linear probe.
No need for alignment or histology. Just the signals and the embedding.
(8/13)
These groups differ in firing rates (awake and sleep), theta modulation, and coupling to gamma and ripple.
This let us replicate findings from fancy silicon probes using tetrodes, and extend them further.
(7/13)
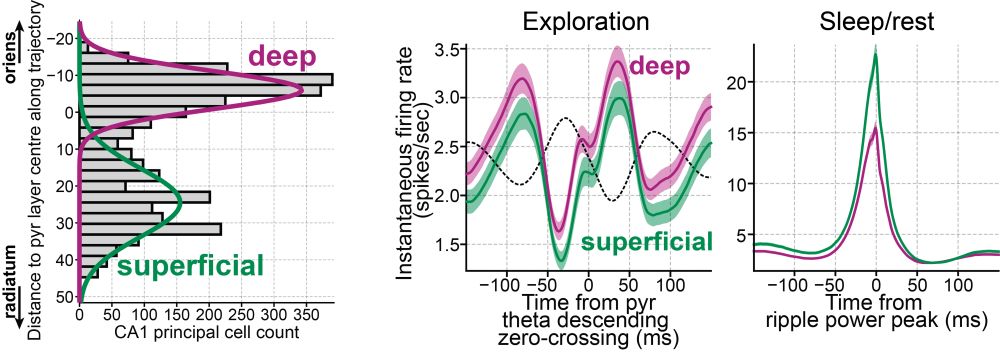
These groups differ in firing rates (awake and sleep), theta modulation, and coupling to gamma and ripple.
This let us replicate findings from fancy silicon probes using tetrodes, and extend them further.
(7/13)
In pyr fast gamma, principal cells fire before interneurons. In rad fast gamma, this flips!
Same freq range, but ≠ mechanism.
+ pyr fast gamma and ripples may arise from the same oscillator: called 'fast gamma' when theta-driven, 'ripples' when driven by SW inputs.
(6/13)

In pyr fast gamma, principal cells fire before interneurons. In rad fast gamma, this flips!
Same freq range, but ≠ mechanism.
+ pyr fast gamma and ripples may arise from the same oscillator: called 'fast gamma' when theta-driven, 'ripples' when driven by SW inputs.
(6/13)
It’s really robust! Ephys patterns do predict anatomical layers.
See two tetrodes going to DG molecular layer below (more examples in the paper!).
(5/13)

It’s really robust! Ephys patterns do predict anatomical layers.
See two tetrodes going to DG molecular layer below (more examples in the paper!).
(5/13)
This spatiotemporal structure was robust across mice in our silicon probe dataset.
See examples below and check the paper for all the details (including a few novel gamma rhythms)!
(3/13)
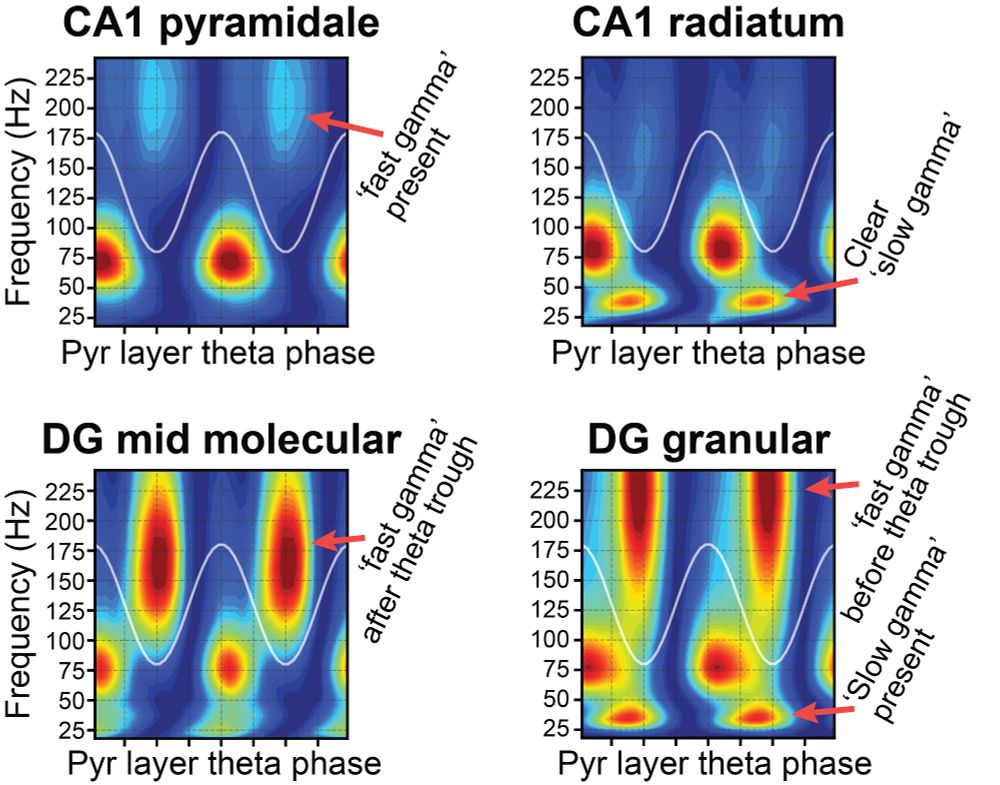
This spatiotemporal structure was robust across mice in our silicon probe dataset.
See examples below and check the paper for all the details (including a few novel gamma rhythms)!
(3/13)
In essence, the embedding links anatomical position to electrophysiological features.
(2/13)

In essence, the embedding links anatomical position to electrophysiological features.
(2/13)
With Demi Brizee & David Dupret, we track how oscillations and spiking behaviour map onto hippocampal layers using an LFP-based embedding.
(1/13)
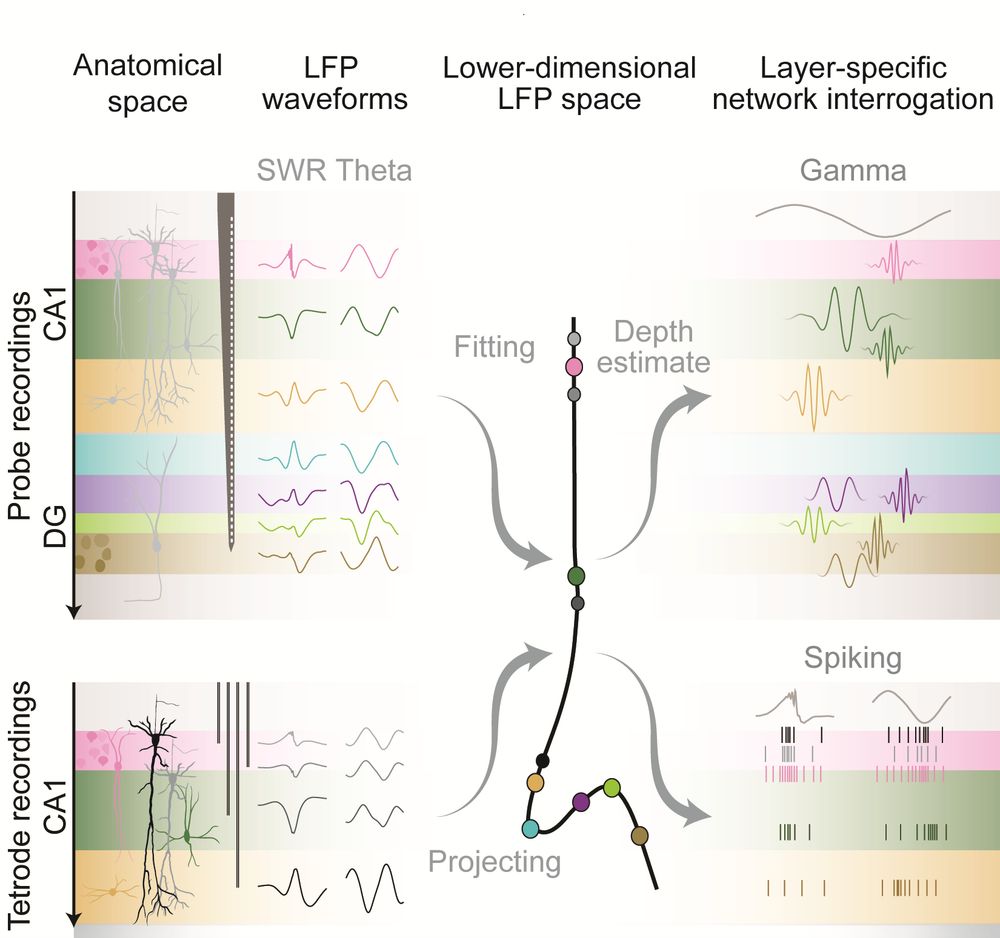
With Demi Brizee & David Dupret, we track how oscillations and spiking behaviour map onto hippocampal layers using an LFP-based embedding.
(1/13)

