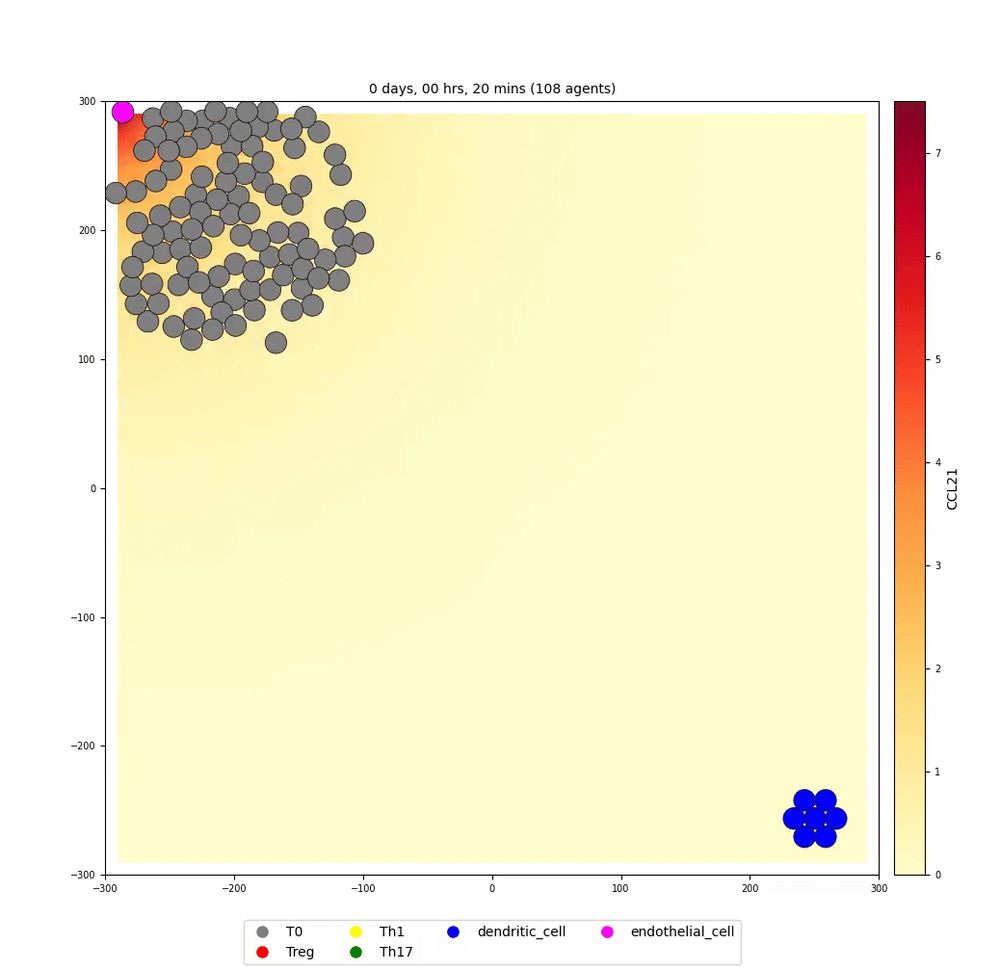
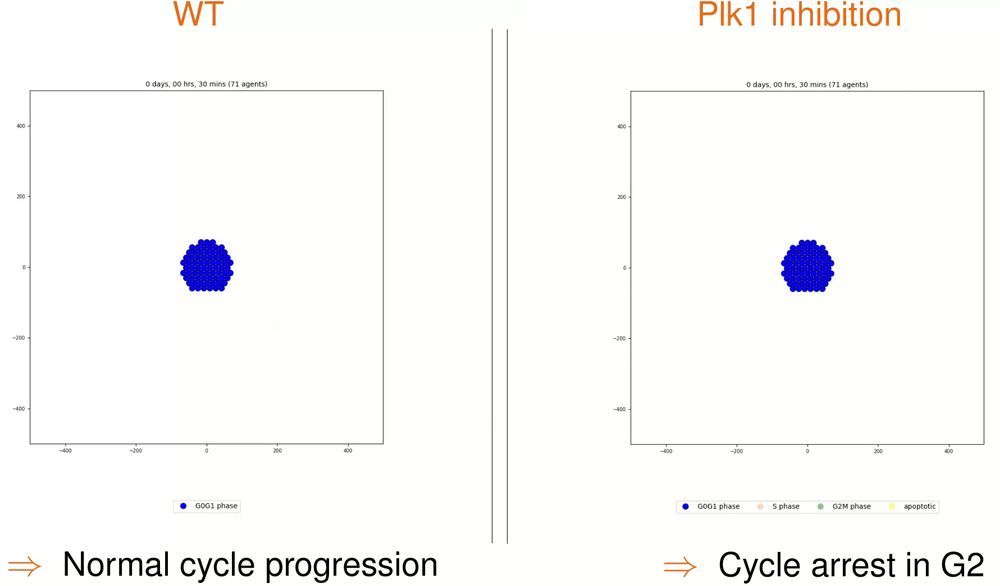
Modeling and simulating biological systems with multiscale frameworks.
#MaBoSS, #PhysiBoSS developer
cancer invasion model, removing most of the C++ code, and replacing it with the new mapping system. A larger scale, real-world example of how to use the latest #PhysiBoSS functionalities!
cancer invasion model, removing most of the C++ code, and replacing it with the new mapping system. A larger scale, real-world example of how to use the latest #PhysiBoSS functionalities!


cancer invasion model, removing most of the C++ code, and replacing it with the new mapping system. A larger scale, real-world example of how to use the latest #PhysiBoSS functionalities!

cancer invasion model, removing most of the C++ code, and replacing it with the new mapping system. A larger scale, real-world example of how to use the latest #PhysiBoSS functionalities!