Valentina Baderna
@valentinabaderna.bsky.social
PhD student @EMBL in the Krebs Lab
Finally, our model predicts that only the endogenous loci should be affected by loss of H3K27Ac as the ectopic landing pad is located within a chromatin neutral environment. And indeed, CREs like the Xpa enhancer lose accessibility endogenously but not when inserted at the ectopic site. 11/11

April 8, 2025 at 1:52 PM
Finally, our model predicts that only the endogenous loci should be affected by loss of H3K27Ac as the ectopic landing pad is located within a chromatin neutral environment. And indeed, CREs like the Xpa enhancer lose accessibility endogenously but not when inserted at the ectopic site. 11/11
To test this, we chemically inhibited p300, a histone acetyltransferase that deposits H3K27Ac at a subset of enhancers in mESCs. As predicted, chromatin accessibility dropped, and the amplitude of the drop scaled with the degree of H3K27Ac reduction. 10/11

April 8, 2025 at 1:52 PM
To test this, we chemically inhibited p300, a histone acetyltransferase that deposits H3K27Ac at a subset of enhancers in mESCs. As predicted, chromatin accessibility dropped, and the amplitude of the drop scaled with the degree of H3K27Ac reduction. 10/11
When we stratified endogenous CREs by their H3K27Ac ChIP-seq signal, we found that the higher the acetylation, the more frequently the chromatin is accessible.
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11

April 8, 2025 at 1:52 PM
When we stratified endogenous CREs by their H3K27Ac ChIP-seq signal, we found that the higher the acetylation, the more frequently the chromatin is accessible.
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
Can transcription factors still find and bind their motif at this ectopic site?
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11

April 8, 2025 at 1:52 PM
Can transcription factors still find and bind their motif at this ectopic site?
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
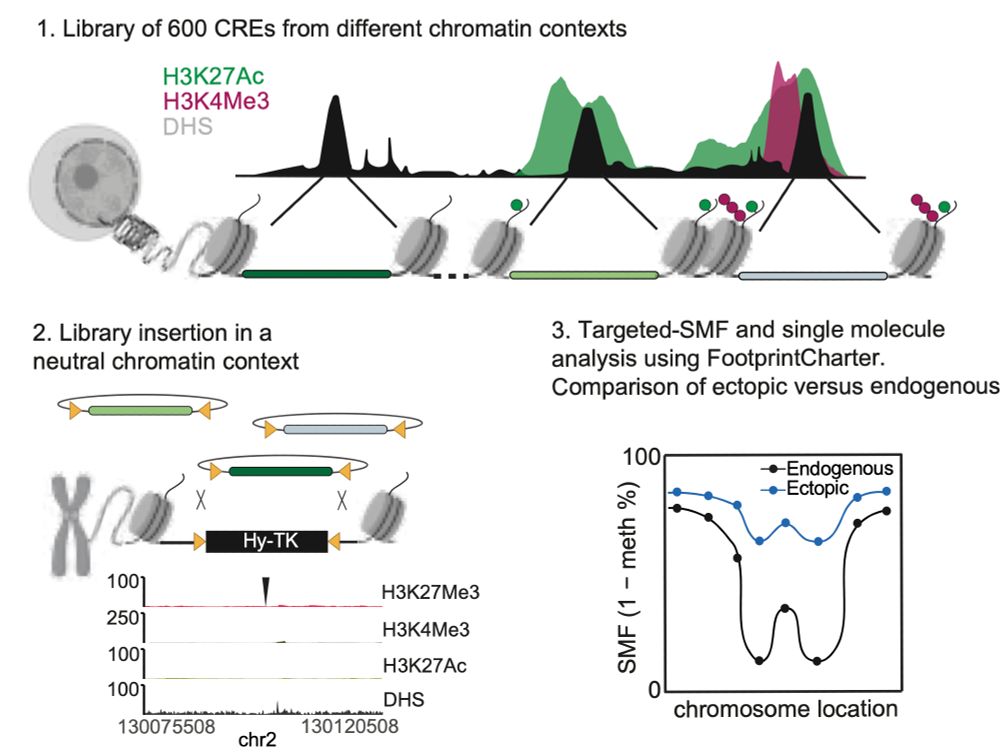
To dissect the contribution of TF binding vs. chromatin context in driving chromatin opening, we developed a Multiplexed Chromatin-Integrated Reporter Assay. It measures the ability of hundreds of CREs (<300 bp) to recruit TFs and establish accessibility at a defined ectopic locus. 7/11
April 8, 2025 at 1:52 PM
To dissect the contribution of TF binding vs. chromatin context in driving chromatin opening, we developed a Multiplexed Chromatin-Integrated Reporter Assay. It measures the ability of hundreds of CREs (<300 bp) to recruit TFs and establish accessibility at a defined ectopic locus. 7/11
We tested this idea by perturbing TF binding. At the motif level, we leveraged the natural genetic variation between mice species crossed into F1 lines. Here, SNPs reduced TF motif affinity across alleles. CREs losing single TF binding showed <10% reductions in chromatin accessibility frequency.6/11

April 8, 2025 at 1:52 PM
We tested this idea by perturbing TF binding. At the motif level, we leveraged the natural genetic variation between mice species crossed into F1 lines. Here, SNPs reduced TF motif affinity across alleles. CREs losing single TF binding showed <10% reductions in chromatin accessibility frequency.6/11
We found that the frequency of chromatin accessibility at CREs scales with their increasing number of bound TF motifs. Overall, any additional TF binding event increases chromatin accessibility frequency and width rather moderately, i.e., by ~10%. 5/11

April 8, 2025 at 1:52 PM
We found that the frequency of chromatin accessibility at CREs scales with their increasing number of bound TF motifs. Overall, any additional TF binding event increases chromatin accessibility frequency and width rather moderately, i.e., by ~10%. 5/11
We quantified the heterogeneity of chromatin accessibility at mouse ESC CREs. We observed that, within the same cell population, accessible molecules systematically co-exist with nucleosome-occupied ones. At enhancers this is much more prominent than at promoters. 4/11

April 8, 2025 at 1:52 PM
We quantified the heterogeneity of chromatin accessibility at mouse ESC CREs. We observed that, within the same cell population, accessible molecules systematically co-exist with nucleosome-occupied ones. At enhancers this is much more prominent than at promoters. 4/11
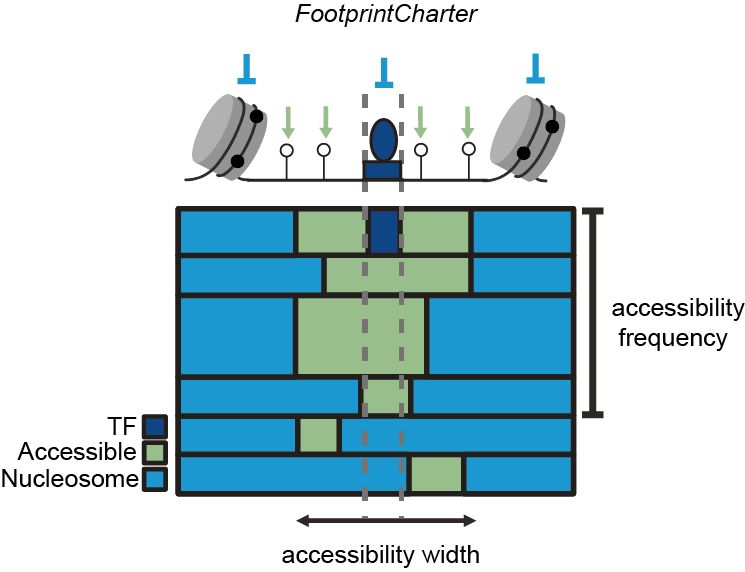
We developed FootprintCharter to classify molecules into accessible or nucleosome-occupied at TF motifs by unsupervisedly detecting TF and nucleosome footprints. FootprintCharter is distributed on Bioconductor at bit.ly/3XLe8RC. For more details, read our annex preprint at bit.ly/3EkRqJh. 3/11
April 8, 2025 at 1:52 PM
We developed FootprintCharter to classify molecules into accessible or nucleosome-occupied at TF motifs by unsupervisedly detecting TF and nucleosome footprints. FootprintCharter is distributed on Bioconductor at bit.ly/3XLe8RC. For more details, read our annex preprint at bit.ly/3EkRqJh. 3/11
Bulk assays such as ATAC-seq selectively sequence accessible loci over nucleosome-occupied ones, losing information on accessibility frequency at CREs. Instead, SMF marks accessibility by methylation footprinting and sequences all molecules, revealing the frequency of accessibility across cells.2/11

April 8, 2025 at 1:52 PM
Bulk assays such as ATAC-seq selectively sequence accessible loci over nucleosome-occupied ones, losing information on accessibility frequency at CREs. Instead, SMF marks accessibility by methylation footprinting and sequences all molecules, revealing the frequency of accessibility across cells.2/11
Finally, our model predicts that only the endogenous loci should be affected by loss of H3K27Ac as the ectopic landing pad is located within a chromatin neutral environment. And indeed, CREs like the Xpa enhancer lose accessibility endogenously but not when inserted at the ectopic site. 11/11

April 8, 2025 at 1:22 PM
Finally, our model predicts that only the endogenous loci should be affected by loss of H3K27Ac as the ectopic landing pad is located within a chromatin neutral environment. And indeed, CREs like the Xpa enhancer lose accessibility endogenously but not when inserted at the ectopic site. 11/11
To test this, we chemically inhibited p300, a histone acetyltransferase that deposits H3K27Ac at a subset of enhancers in mESCs. As predicted, chromatin accessibility dropped, and the amplitude of the drop scaled with the degree of H3K27Ac reduction. 10/11

April 8, 2025 at 1:22 PM
To test this, we chemically inhibited p300, a histone acetyltransferase that deposits H3K27Ac at a subset of enhancers in mESCs. As predicted, chromatin accessibility dropped, and the amplitude of the drop scaled with the degree of H3K27Ac reduction. 10/11
When we stratified endogenous CREs by their H3K27Ac ChIP-seq signal, we found that the higher the acetylation, the more frequently the chromatin is accessible.
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11

April 8, 2025 at 1:22 PM
When we stratified endogenous CREs by their H3K27Ac ChIP-seq signal, we found that the higher the acetylation, the more frequently the chromatin is accessible.
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
Can transcription factors still find and bind their motif at this ectopic site?
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11

April 8, 2025 at 1:22 PM
Can transcription factors still find and bind their motif at this ectopic site?
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
To dissect the contribution of TF binding vs. chromatin context in driving chromatin opening, we developed a Multiplexed Chromatin-Integrated Reporter Assay. It measures the ability of hundreds of CREs (<300 bp) to recruit TFs and establish accessibility at a defined neutral ectopic locus. 7/11

April 8, 2025 at 1:22 PM
To dissect the contribution of TF binding vs. chromatin context in driving chromatin opening, we developed a Multiplexed Chromatin-Integrated Reporter Assay. It measures the ability of hundreds of CREs (<300 bp) to recruit TFs and establish accessibility at a defined neutral ectopic locus. 7/11
We tested this idea by perturbing TF binding. At the motif level, we leveraged the natural genetic variation between mice species crossed into F1 lines. Here, SNPs reduced TF motif affinity across alleles. CREs losing single TF binding, showed<10% reductions in chromatin accessibility frequency.6/11

April 8, 2025 at 1:22 PM
We tested this idea by perturbing TF binding. At the motif level, we leveraged the natural genetic variation between mice species crossed into F1 lines. Here, SNPs reduced TF motif affinity across alleles. CREs losing single TF binding, showed<10% reductions in chromatin accessibility frequency.6/11
We found that the frequency of chromatin accessibility at CREs scales with their increasing number of bound TF motifs. Overall, any additional TF binding event increases chromatin accessibility frequency and width rather moderately, i.e., by ~10%. 5/11

April 8, 2025 at 1:22 PM
We found that the frequency of chromatin accessibility at CREs scales with their increasing number of bound TF motifs. Overall, any additional TF binding event increases chromatin accessibility frequency and width rather moderately, i.e., by ~10%. 5/11
We quantified the heterogeneity of chromatin accessibility at mouse ESC CREs. We observed that, within the same cell population, accessible molecules systematically co-exist with nucleosome-occupied ones. At enhancers this is much more prominent than at promoters. 4/11

April 8, 2025 at 1:22 PM
We quantified the heterogeneity of chromatin accessibility at mouse ESC CREs. We observed that, within the same cell population, accessible molecules systematically co-exist with nucleosome-occupied ones. At enhancers this is much more prominent than at promoters. 4/11
We developed FootprintCharter to classify molecules into accessible or nucleosome-occupied at TF motifs by unsupervisedly detecting TF and nucleosome footprints. FootprintCharter is distributed on Bioconductor at bit.ly/3XLe8RC. For more details, read our annex preprint at bit.ly/3EkRqJh. 3/11

April 8, 2025 at 1:22 PM
We developed FootprintCharter to classify molecules into accessible or nucleosome-occupied at TF motifs by unsupervisedly detecting TF and nucleosome footprints. FootprintCharter is distributed on Bioconductor at bit.ly/3XLe8RC. For more details, read our annex preprint at bit.ly/3EkRqJh. 3/11
Bulk assays such as ATAC-seq selectively sequence accessible loci over nucleosome-occupied ones, losing information on accessibility frequency at CREs. Instead, SMF marks accessibility by methylation footprinting and sequences all molecules, revealing the frequency of accessibility across cells.2/11

April 8, 2025 at 1:22 PM
Bulk assays such as ATAC-seq selectively sequence accessible loci over nucleosome-occupied ones, losing information on accessibility frequency at CREs. Instead, SMF marks accessibility by methylation footprinting and sequences all molecules, revealing the frequency of accessibility across cells.2/11
When we stratified endogenous CREs by their H3K27Ac ChIP-seq signal, we found that the higher the acetylation, the more frequently the chromatin is accessible.
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11

April 8, 2025 at 12:55 PM
When we stratified endogenous CREs by their H3K27Ac ChIP-seq signal, we found that the higher the acetylation, the more frequently the chromatin is accessible.
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
But at the ectopic site, this scaling disappears—suggesting that H3K27Ac enhances accessibility frequency endogenously. 9/11
Can transcription factors still find and bind their motif at this ectopic site?
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11

April 8, 2025 at 12:55 PM
Can transcription factors still find and bind their motif at this ectopic site?
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
Yes! But while they do bind, they typically open chromatin to a lower extent than at their endogenous counterparts—with the notable exception of the insulator CTCF. 8/11
To dissect the contribution of TF binding vs. chromatin context in driving chromatin opening, we developed a Multiplexed Chromatin-Integrated Reporter Assay. It measures the ability of hundreds of CREs (<300 bp) to recruit TFs and establish accessibility at a defined neutral ectopic locus. 7/11

April 8, 2025 at 12:55 PM
To dissect the contribution of TF binding vs. chromatin context in driving chromatin opening, we developed a Multiplexed Chromatin-Integrated Reporter Assay. It measures the ability of hundreds of CREs (<300 bp) to recruit TFs and establish accessibility at a defined neutral ectopic locus. 7/11
We tested this idea by perturbing TF binding. At the motif level, we leveraged the natural genetic variation between mice species crossed into F1 lines. Here, SNPs reduced TF motif affinity across alleles. CREs losing single TF binding showed<10% reductions in chromatin accessibility frequency. 6/11

April 8, 2025 at 12:55 PM
We tested this idea by perturbing TF binding. At the motif level, we leveraged the natural genetic variation between mice species crossed into F1 lines. Here, SNPs reduced TF motif affinity across alleles. CREs losing single TF binding showed<10% reductions in chromatin accessibility frequency. 6/11

