
From conda to plugins to Jupyter, practicals on real data with expert guidance.
For Python/ImageJ coders. Apply by 1 Sept – limited to 30! crick.ac.uk/whats-on/cbias-napari-workshop-2025

From conda to plugins to Jupyter, practicals on real data with expert guidance.
For Python/ImageJ coders. Apply by 1 Sept – limited to 30! crick.ac.uk/whats-on/cbias-napari-workshop-2025
It contains sample data, allows to read CZI metadata and can read CZI images (or parts of it) by using the sliders to crop what should be read.
Still quite some things to improve ...
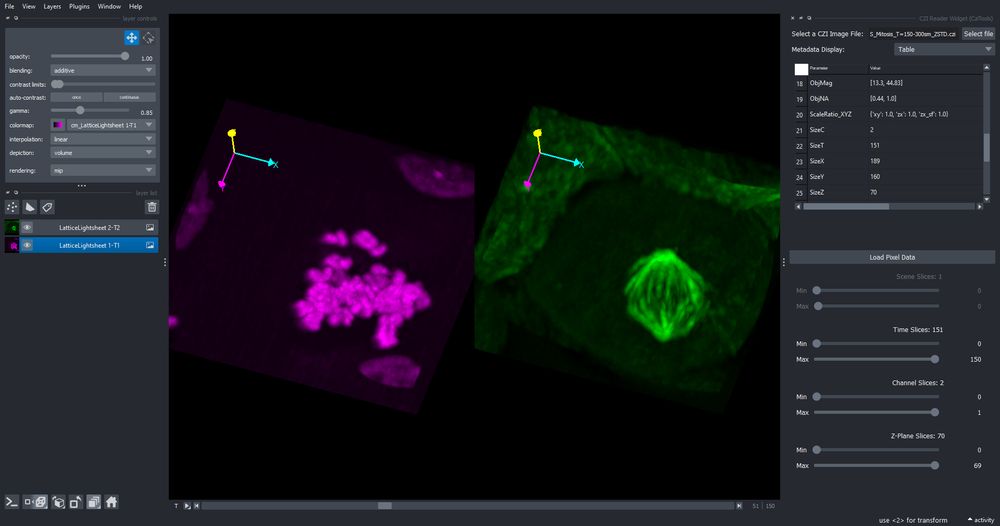
It contains sample data, allows to read CZI metadata and can read CZI images (or parts of it) by using the sliders to crop what should be read.
Still quite some things to improve ...
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.

🌍 www.fabriziomusacchio.com/teaching/tea...
🌍 www.fabriziomusacchio.com/teaching/tea...
💡 SAMJ is a new ImageJ/Fiji plugin using Segment Anything (SAM) for fast, interactive segmentation — no GPU needed, one-click install, real-time results.
🔬 Perfect for large biomedical images.
arxiv.org/abs/2506.02783
#AI #Bioimaging #ImageJ #OpenSource
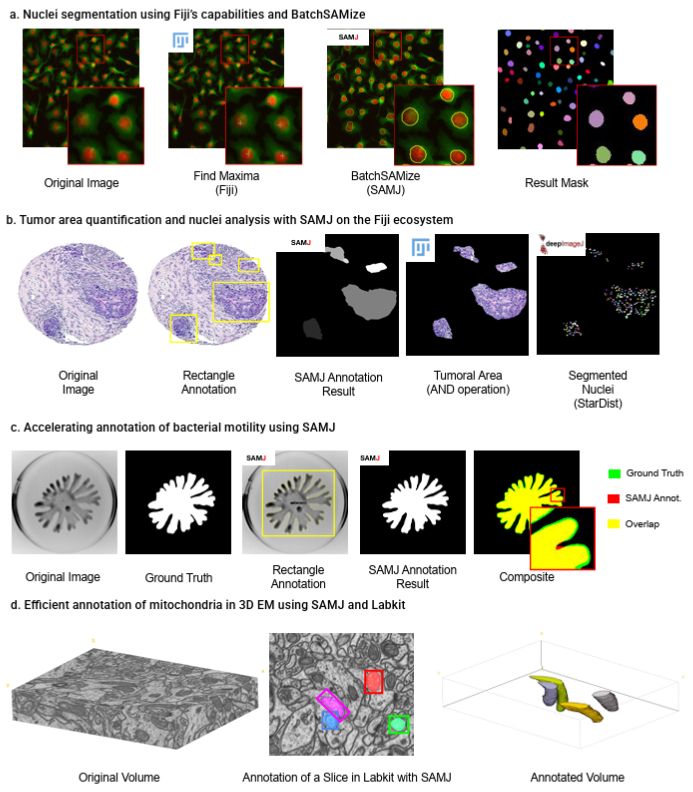
💡 SAMJ is a new ImageJ/Fiji plugin using Segment Anything (SAM) for fast, interactive segmentation — no GPU needed, one-click install, real-time results.
🔬 Perfect for large biomedical images.
arxiv.org/abs/2506.02783
#AI #Bioimaging #ImageJ #OpenSource
- Improvements for automatic tracking.
- A new experimental mode for object classification.
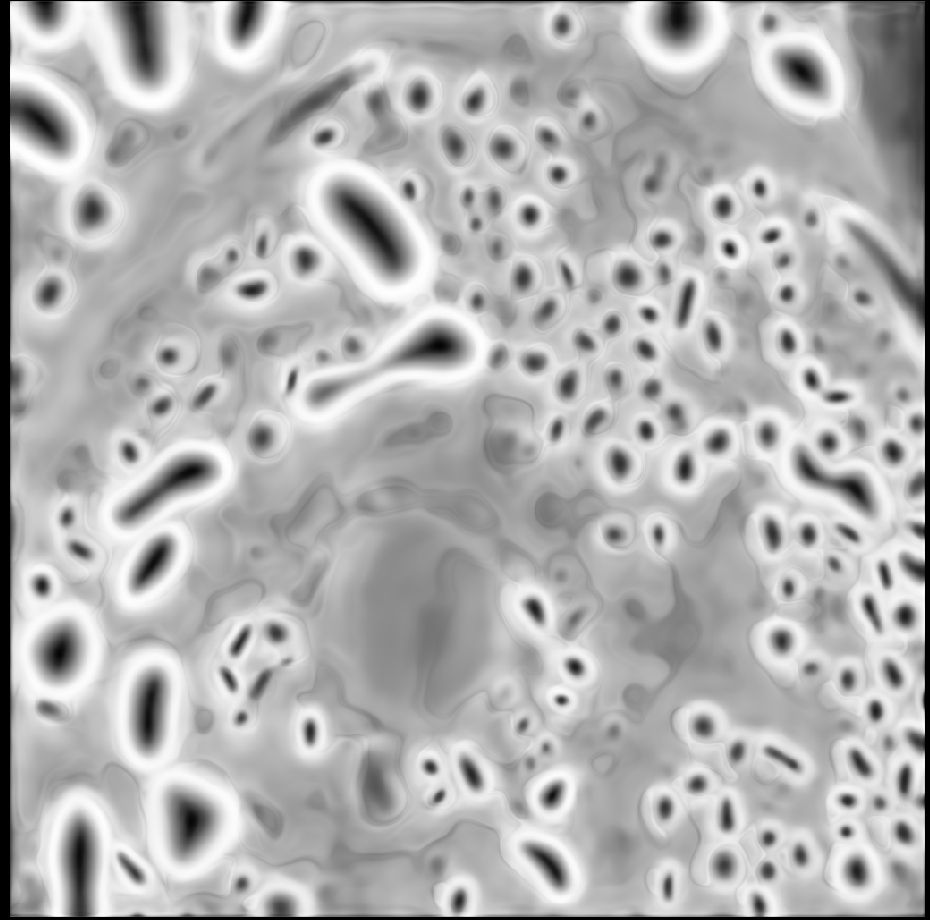
- **New versions of the LM and EM models**
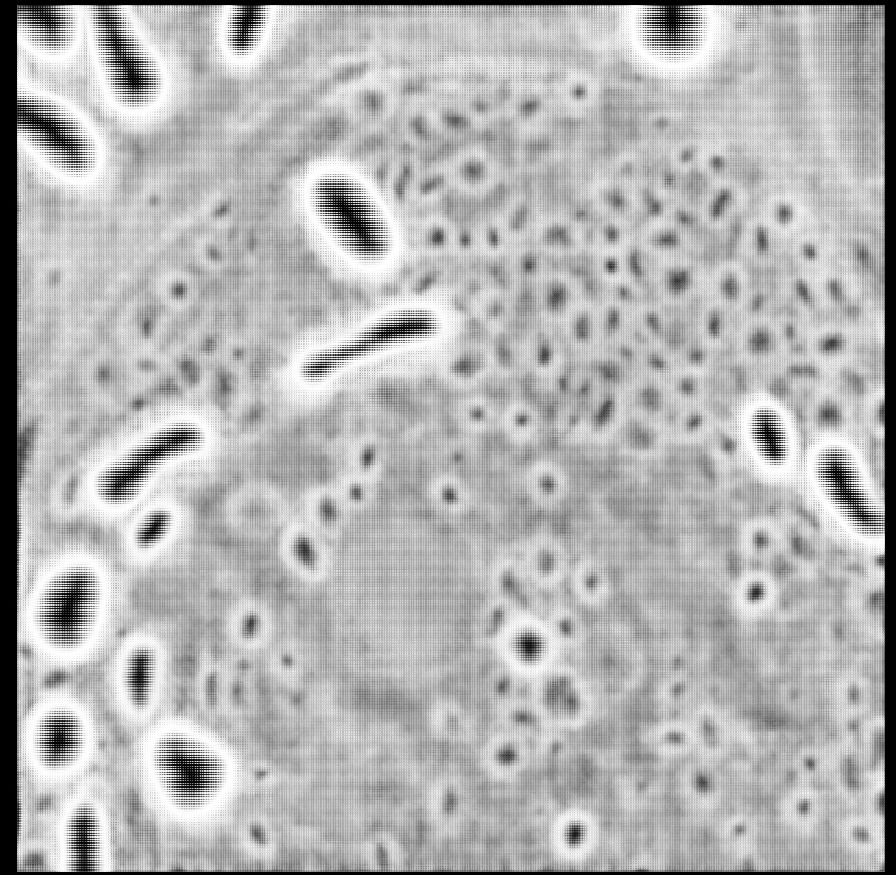
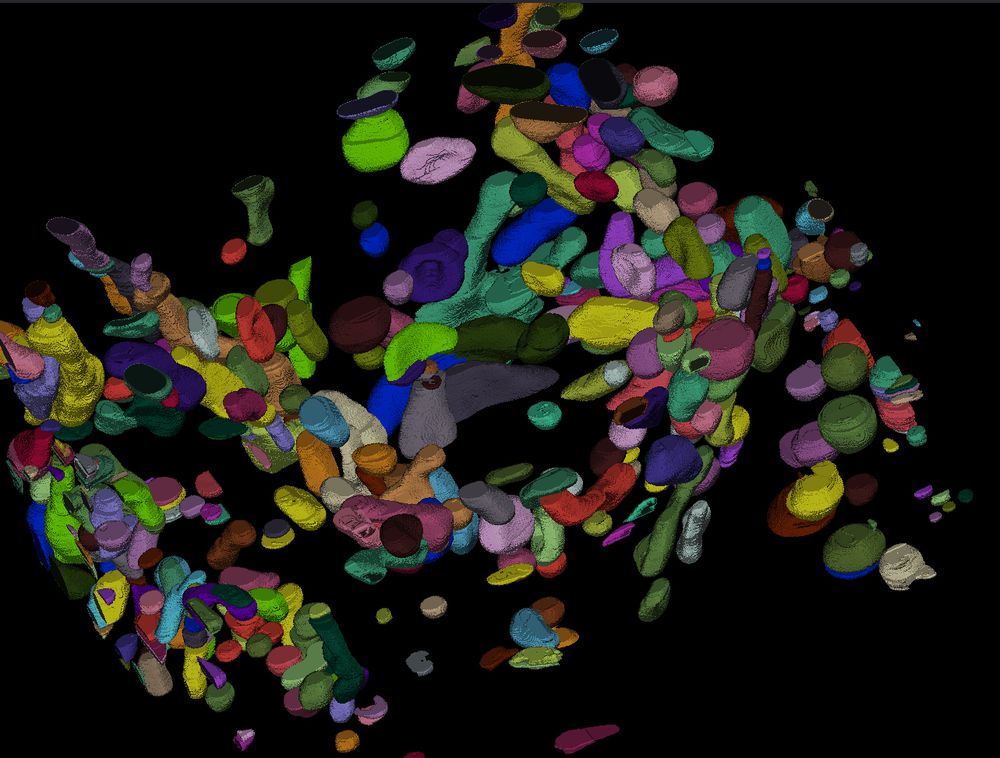
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
- Improvements for automatic tracking.
- A new experimental mode for object classification.
- **New versions of the LM and EM models**
The models fix artifacts in automatic segmentation, see old vs. new prediction and better 3D segmentation results due to it.
Lea Kabjesz shares a step-by-step guide to get you started annotating 2D and 3D images in Cellpose, including video tutorials.
focalplane.biologists.com/2025/06/05/a...
#bioimageanalysis #cellpose #imaging #HowTo

Lea Kabjesz shares a step-by-step guide to get you started annotating 2D and 3D images in Cellpose, including video tutorials.
focalplane.biologists.com/2025/06/05/a...
#bioimageanalysis #cellpose #imaging #HowTo
www.eurobioimaging-access.eu/nodes/uk-node
www.eurobioimaging-access.eu/nodes/uk-node
Enter BioEngine
ai4life.eurobioimaging.eu/announcing-b...
Your go-to for flexible, self-hosted bioimage analysis. Open source, scalable, and built for you!

Enter BioEngine
ai4life.eurobioimaging.eu/announcing-b...
Your go-to for flexible, self-hosted bioimage analysis. Open source, scalable, and built for you!

More details on the conference website.

More details on the conference website.
Here’s a Jupyter notebook I wrote on how, why & when normalization matters, feedback 💛ly welcome.
👇
haesleinhuepf.github.io/BioImageAnal...
github.com/dav1dcg/neur...

github.com/dav1dcg/neur...
𝐃𝐫. 𝐓𝐚𝐥𝐥𝐞𝐲 𝐋𝐚𝐦𝐛𝐞𝐫𝐭 @talley.codes (CITE, Harvard Medical School):
𝐒𝐢𝐦𝐮𝐥𝐚𝐭𝐢𝐧𝐠 𝐫𝐞𝐚𝐥𝐢𝐬𝐭𝐢𝐜 𝐥𝐢𝐠𝐡𝐭 𝐦𝐢𝐜𝐫𝐨𝐬𝐜𝐨𝐩𝐲 𝐢𝐦𝐚𝐠𝐞𝐬 𝐮𝐬𝐢𝐧𝐠 𝐦𝐢𝐜𝐫𝐨𝐬𝐢𝐦
When: 𝐌𝐚𝐲 𝟐𝟗𝐭𝐡 @ 𝟏𝟏 𝐚𝐦 𝐄𝐃𝐓 (Boston time)
Join us on Zoom! harvard.zoom.us/j/9762343464...

𝐃𝐫. 𝐓𝐚𝐥𝐥𝐞𝐲 𝐋𝐚𝐦𝐛𝐞𝐫𝐭 @talley.codes (CITE, Harvard Medical School):
𝐒𝐢𝐦𝐮𝐥𝐚𝐭𝐢𝐧𝐠 𝐫𝐞𝐚𝐥𝐢𝐬𝐭𝐢𝐜 𝐥𝐢𝐠𝐡𝐭 𝐦𝐢𝐜𝐫𝐨𝐬𝐜𝐨𝐩𝐲 𝐢𝐦𝐚𝐠𝐞𝐬 𝐮𝐬𝐢𝐧𝐠 𝐦𝐢𝐜𝐫𝐨𝐬𝐢𝐦
When: 𝐌𝐚𝐲 𝟐𝟗𝐭𝐡 @ 𝟏𝟏 𝐚𝐦 𝐄𝐃𝐓 (Boston time)
Join us on Zoom! harvard.zoom.us/j/9762343464...
Get started with Python and learn to analyze fluorescence microscopy images — no coding experience required!
Fee waivers available thanks to funding from @bioimagingna.bsky.social!
🗓️ Apply June 1st!
iac.hms.harvard.edu/bobiac/2025/

Get started with Python and learn to analyze fluorescence microscopy images — no coding experience required!
Fee waivers available thanks to funding from @bioimagingna.bsky.social!
🗓️ Apply June 1st!
iac.hms.harvard.edu/bobiac/2025/
nimbusimage.com

nimbusimage.com


github.com/haesleinhuep...

github.com/haesleinhuep...

