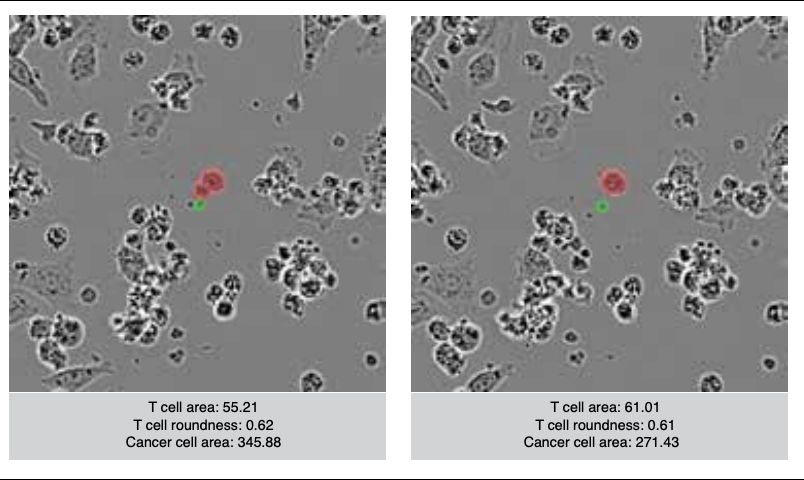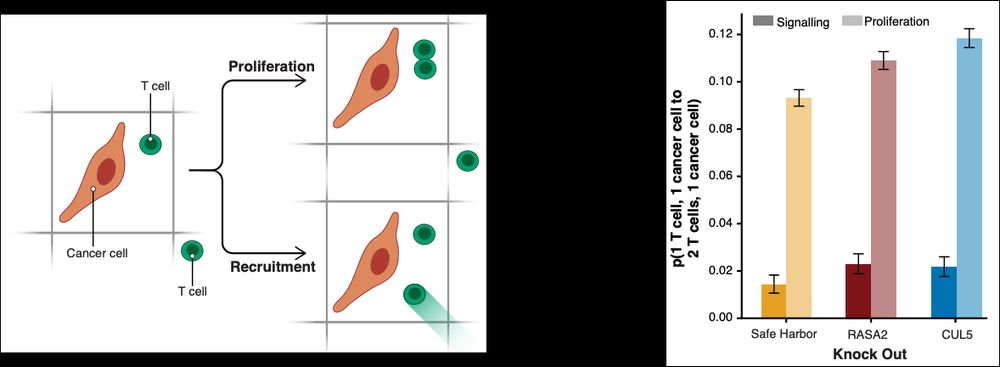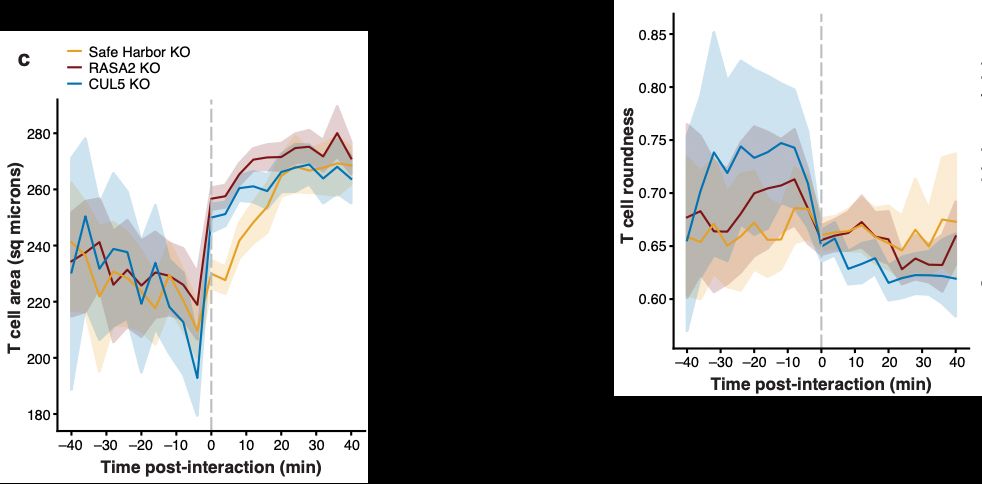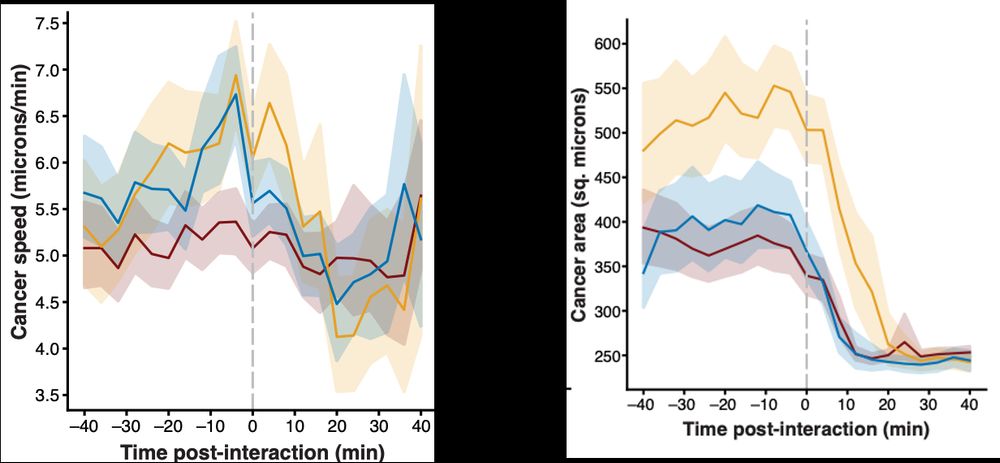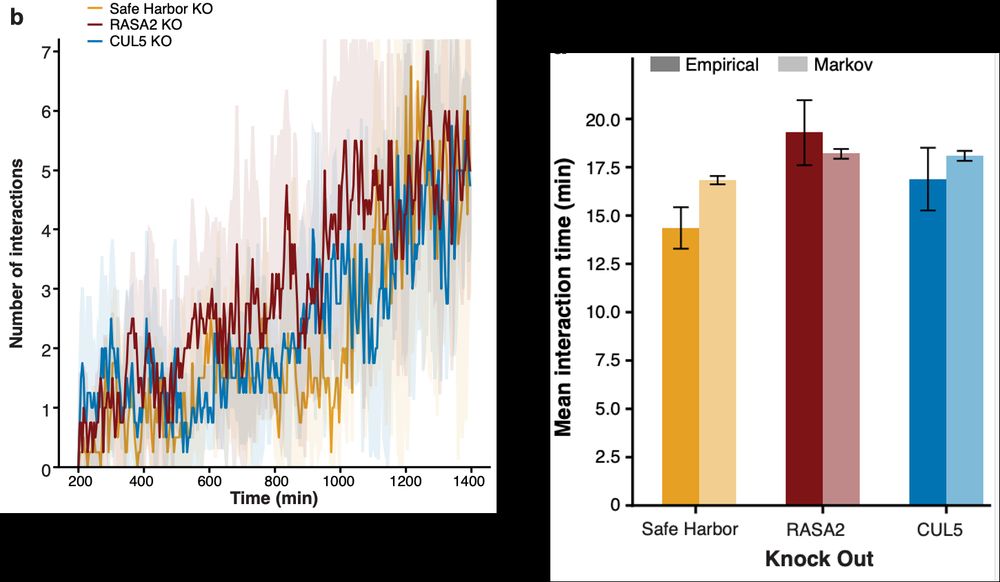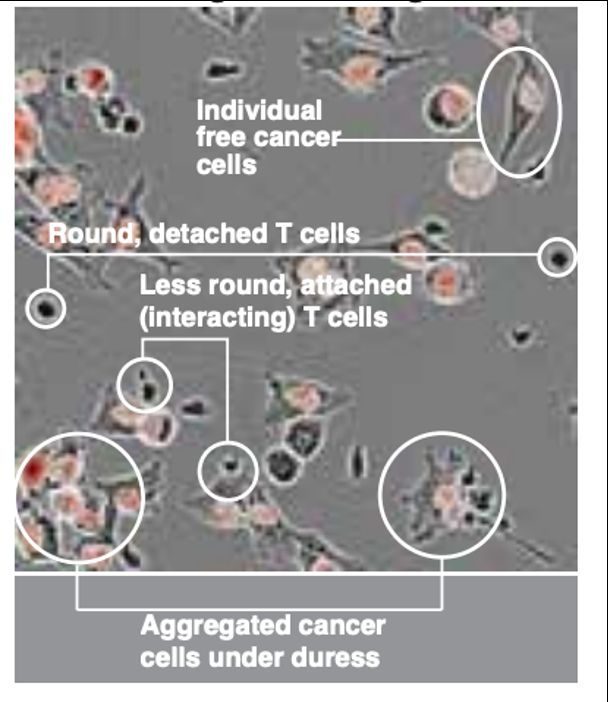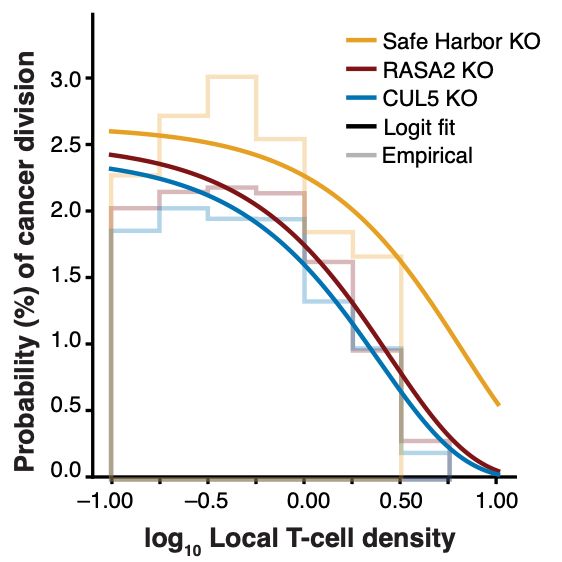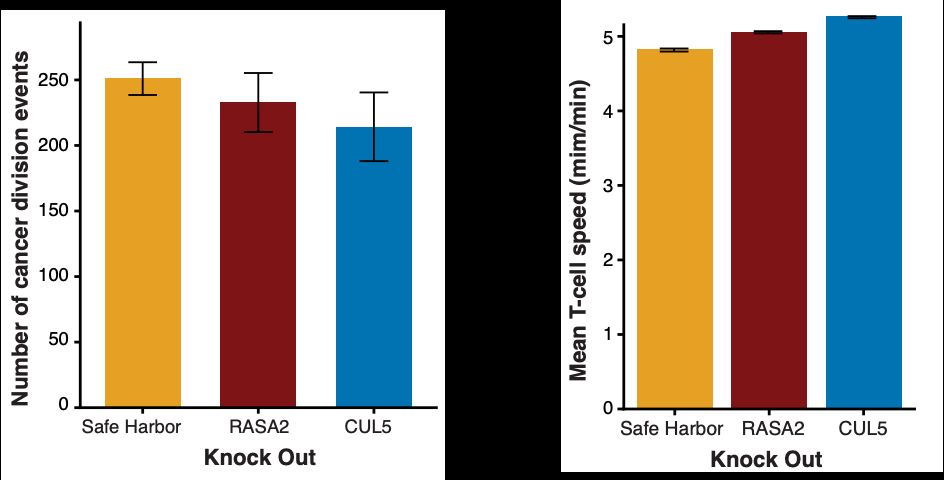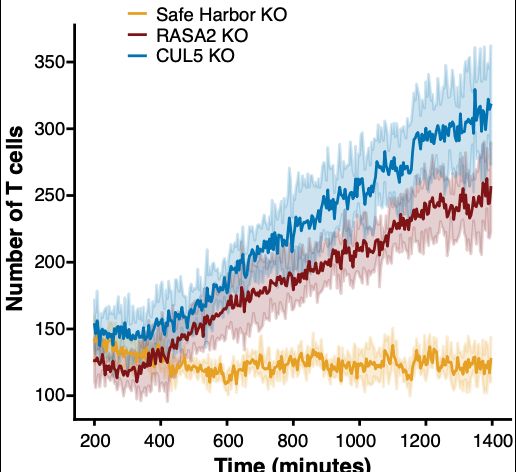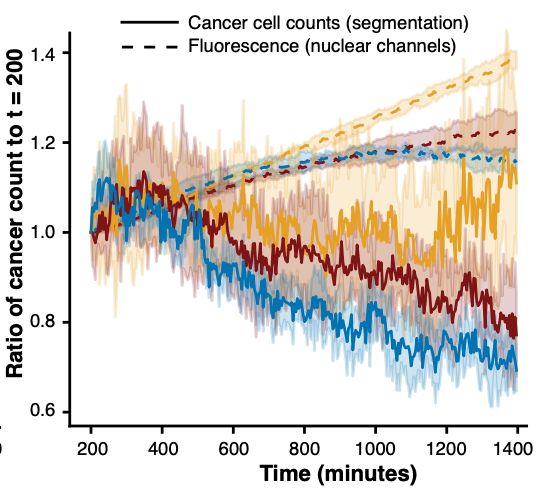
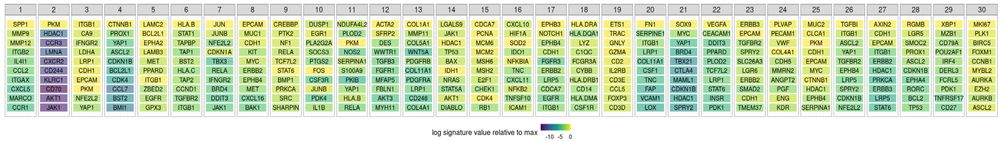
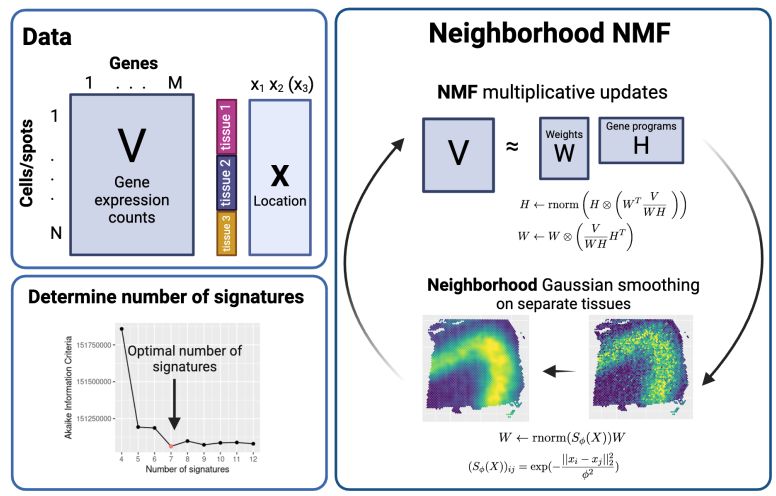
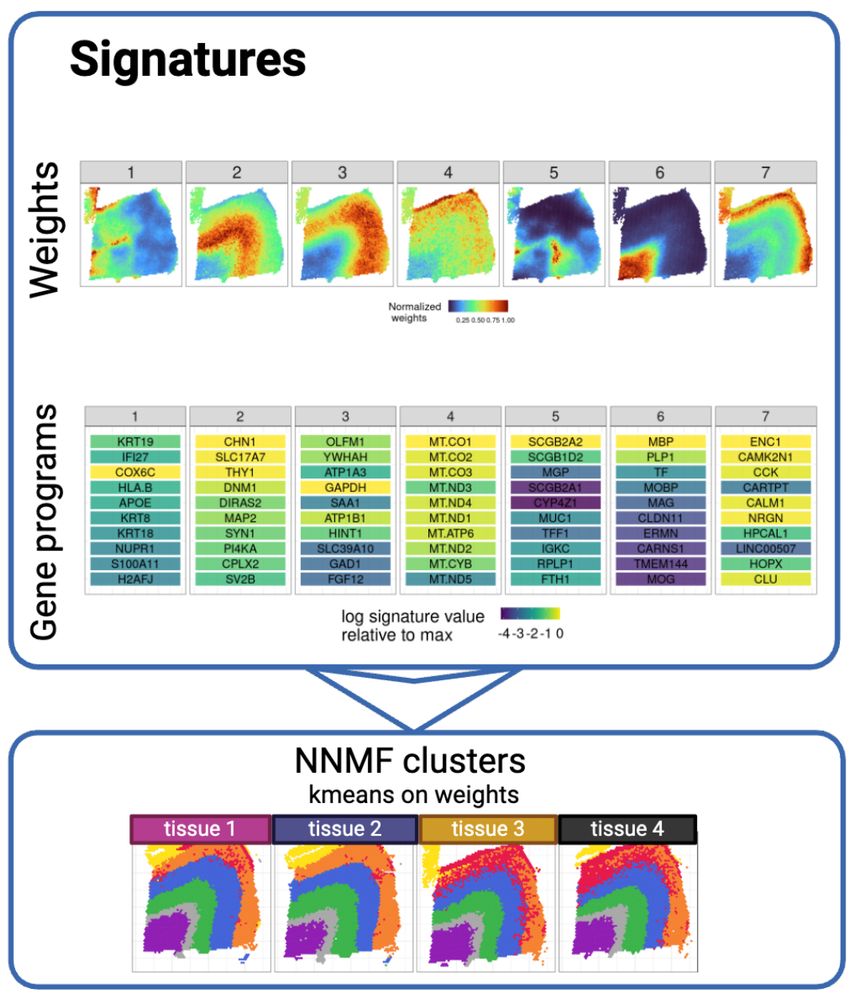
Thank you to @bioimagearchive.bsky.social for hosting these Incucyte image data -- this is a new thing for them, and they have been so kind in working through the details of submission (link coming soon!)! 🎉
Thank you to @bioimagearchive.bsky.social for hosting these Incucyte image data -- this is a new thing for them, and they have been so kind in working through the details of submission (link coming soon!)! 🎉
github.com/vanvalenlab/...
github.com/bee-hive/occ...
github.com/vanvalenlab/...
github.com/bee-hive/occ...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...