We made it easy to:
📊 Add datasets
🖥️ Add new models
📈Benchmark reproducibly
We hope you will give it a try, and if you have any comments or ideas, feel free to reach out!
We made it easy to:
📊 Add datasets
🖥️ Add new models
📈Benchmark reproducibly
We hope you will give it a try, and if you have any comments or ideas, feel free to reach out!
1. genomic sequences != language -> we need training objectives suited to the heterogeneity inherent to genomic data
2. relevant benchmarking tasks and evaluating generalizability is important as these models start to be used for therapeutic design
1. genomic sequences != language -> we need training objectives suited to the heterogeneity inherent to genomic data
2. relevant benchmarking tasks and evaluating generalizability is important as these models start to be used for therapeutic design
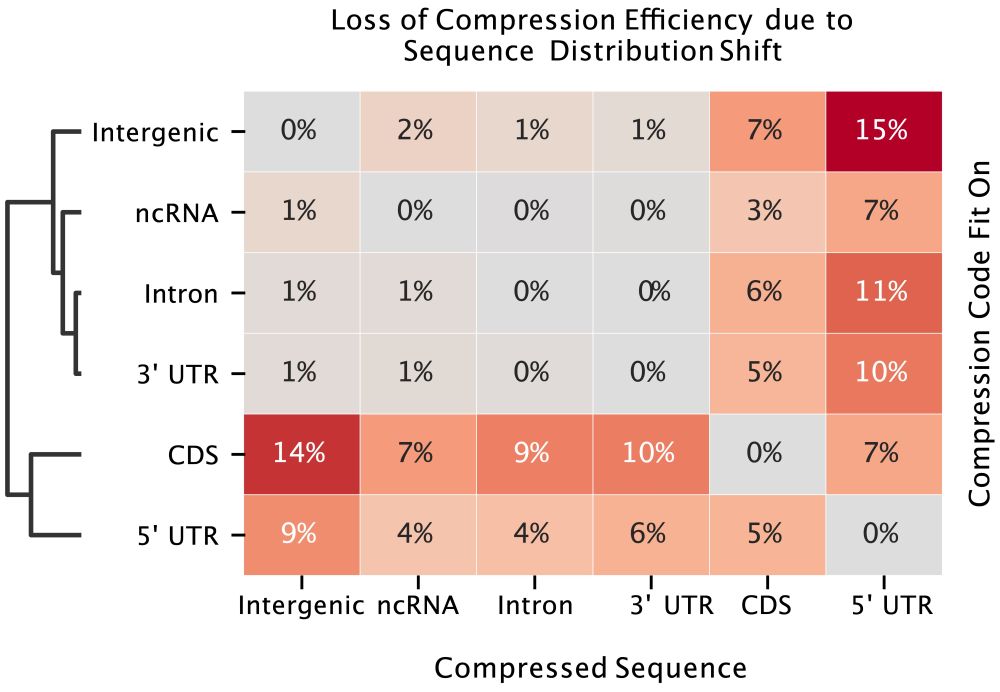
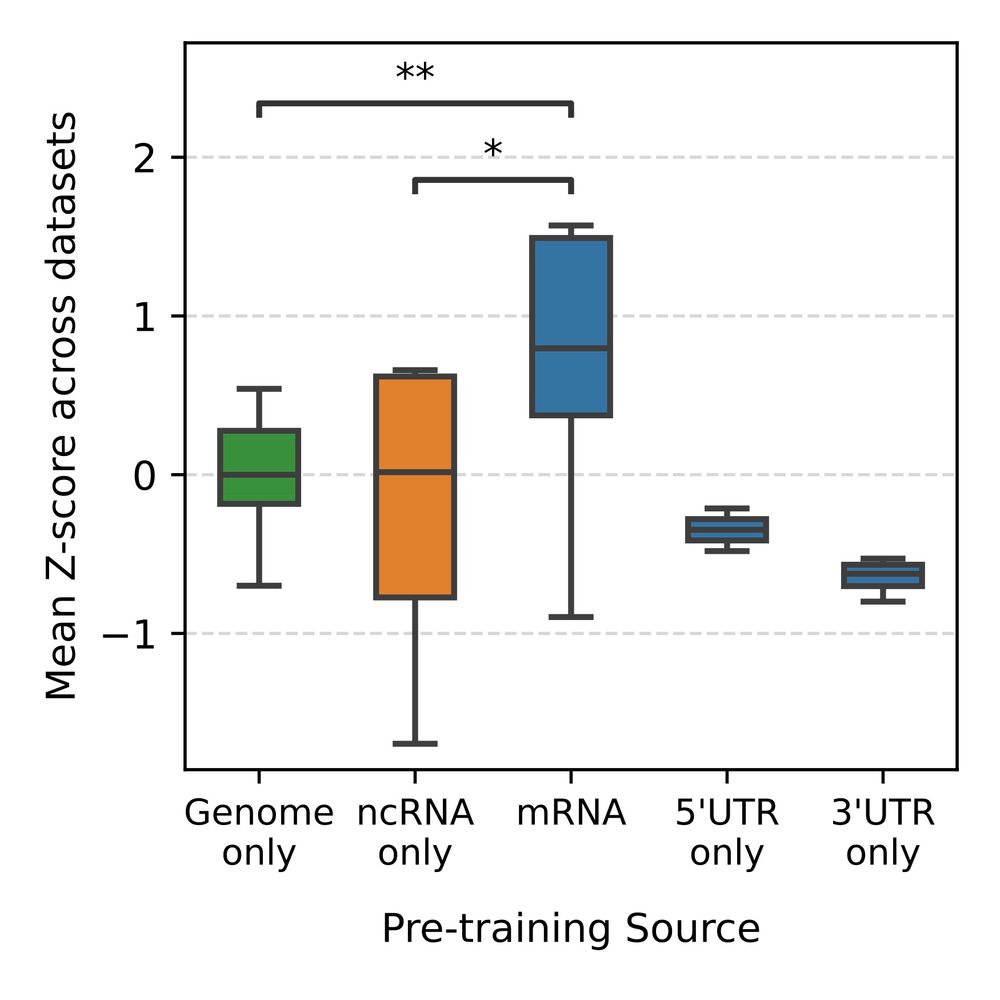
We know that uUAGs reduce translation, and strong Kozak sequences enhance it, so we trained linear probes using 3 subsets of these features and tested on the held out set.
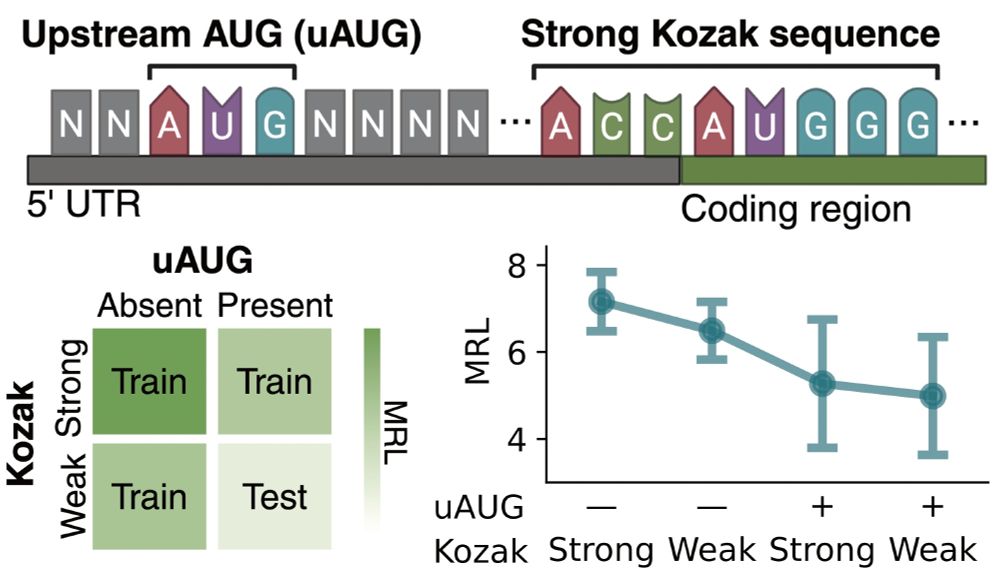
We know that uUAGs reduce translation, and strong Kozak sequences enhance it, so we trained linear probes using 3 subsets of these features and tested on the held out set.
✅ Orthrus+MLM matches/beats SOTA on 6/10 datasets without increasing the training data or significantly increasing model parameters
📈 Pareto-dominates all models larger than 10 million parameters, including Evo2
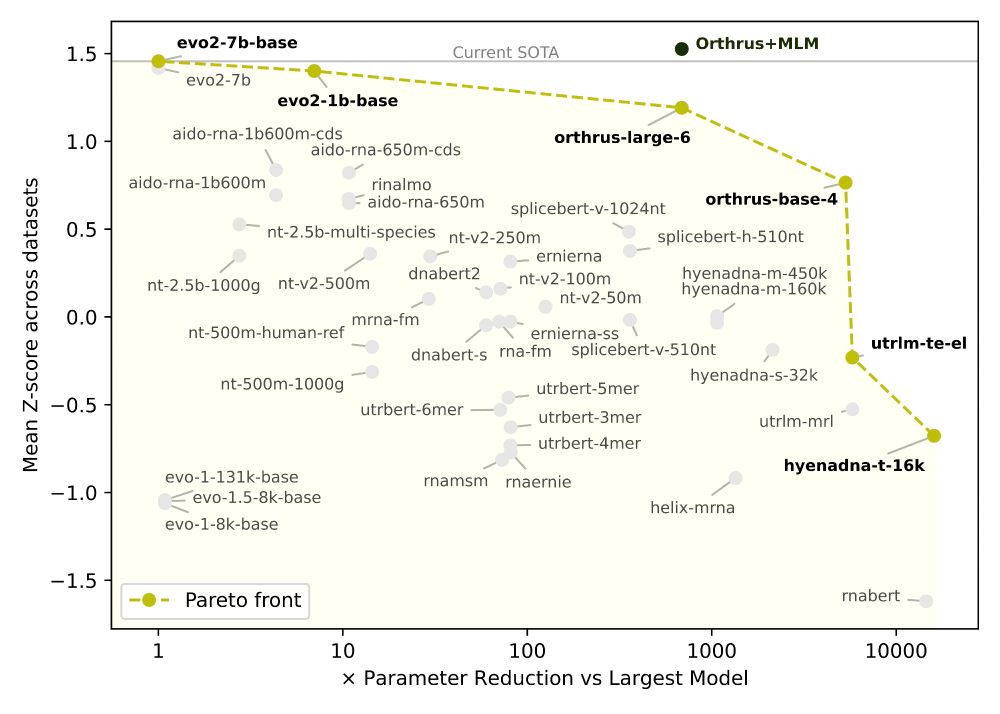
✅ Orthrus+MLM matches/beats SOTA on 6/10 datasets without increasing the training data or significantly increasing model parameters
📈 Pareto-dominates all models larger than 10 million parameters, including Evo2
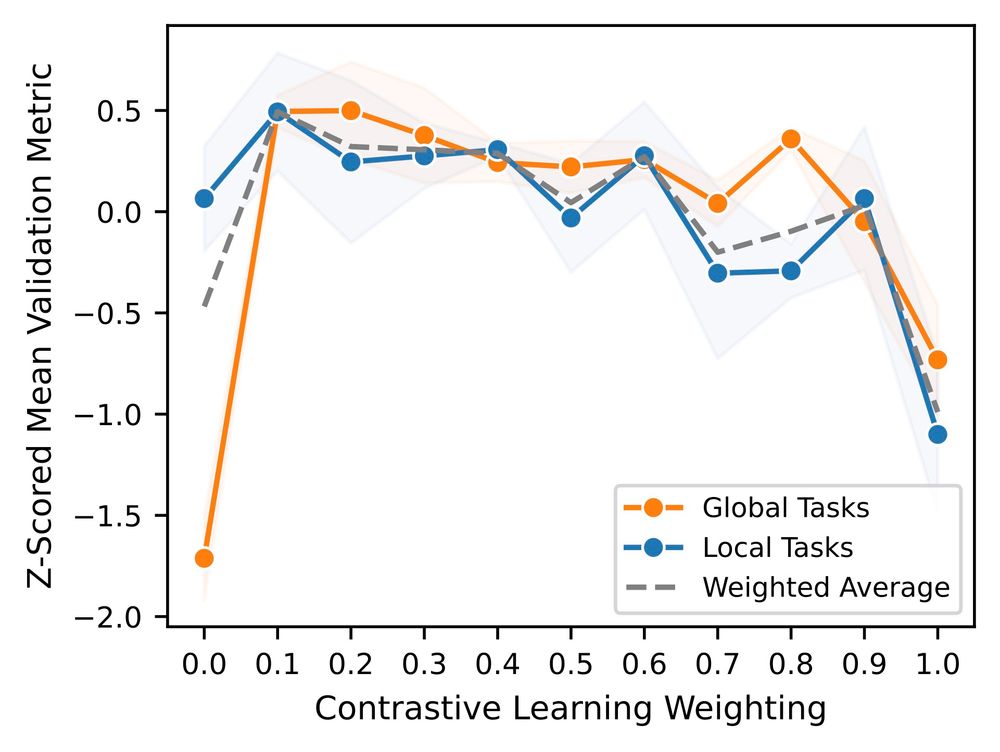
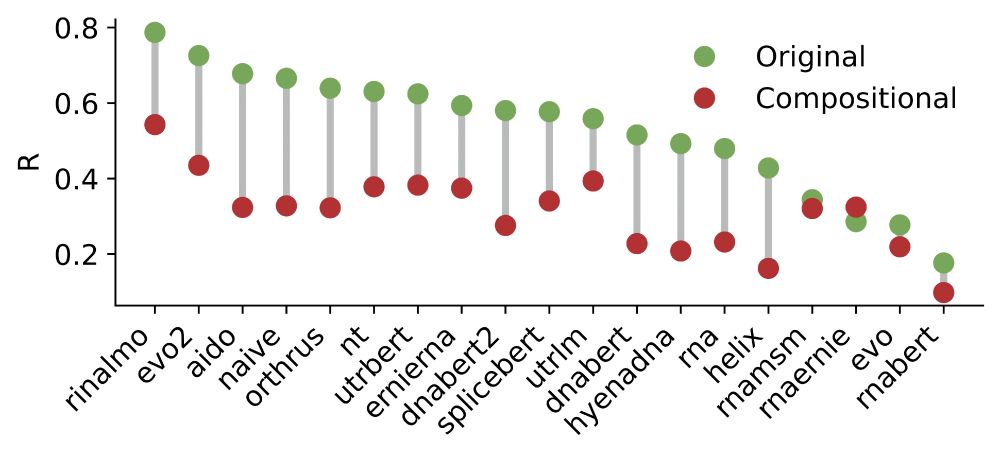
Interestingly, we noticed that the 2nd best model, Orthrus, was not far off compared to Evo2 despite having only 10 million parameters
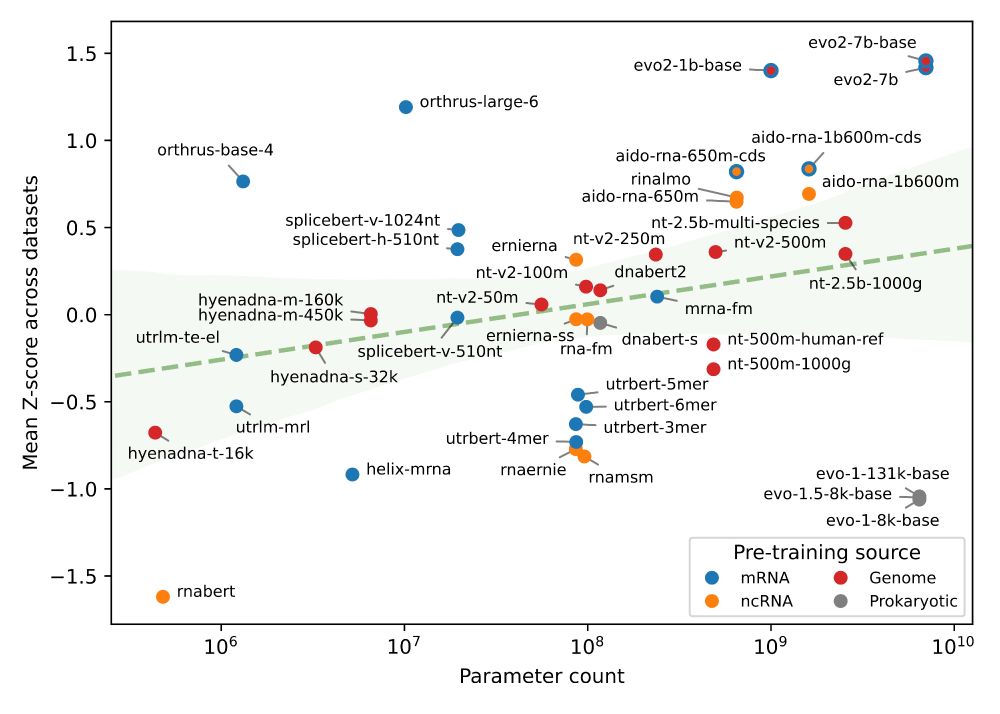
Interestingly, we noticed that the 2nd best model, Orthrus, was not far off compared to Evo2 despite having only 10 million parameters
In total, we conducted over 135,000 total experiments!
So what did we learn? 👇🏽
In total, we conducted over 135,000 total experiments!
So what did we learn? 👇🏽