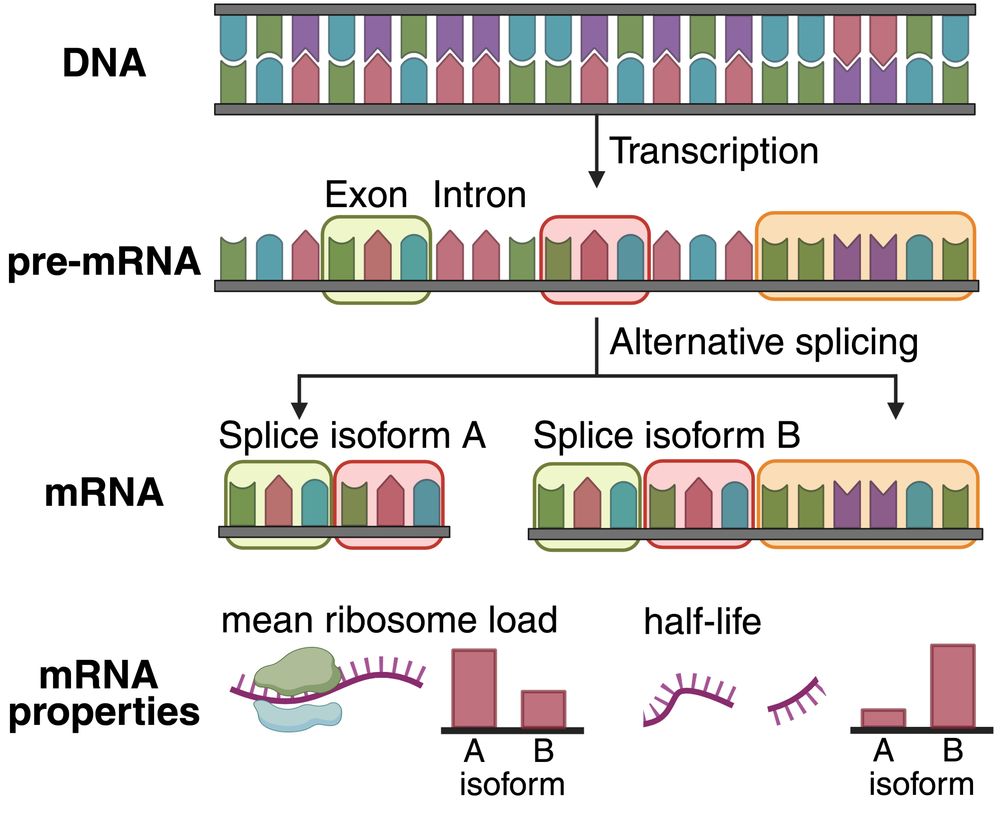
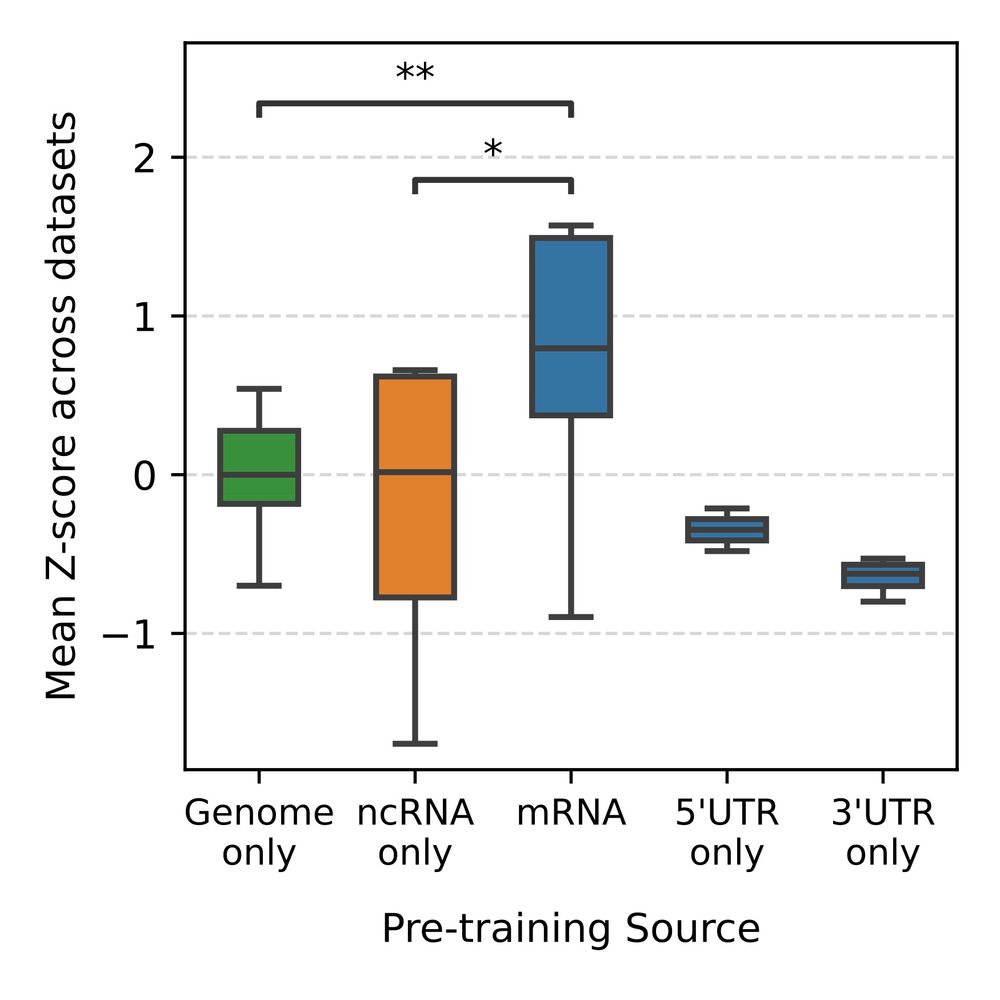
We know that uUAGs reduce translation, and strong Kozak sequences enhance it, so we trained linear probes using 3 subsets of these features and tested on the held out set.
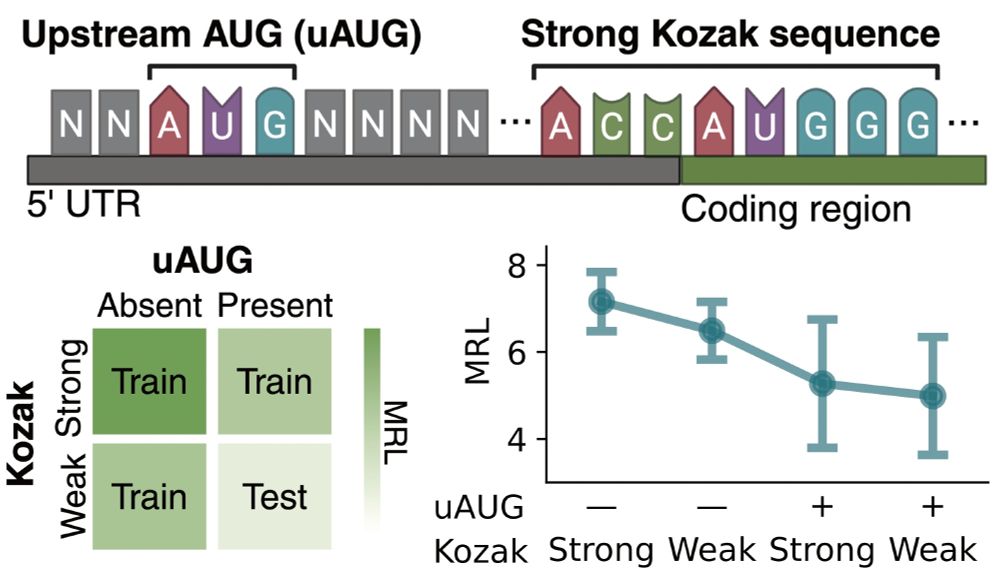
We know that uUAGs reduce translation, and strong Kozak sequences enhance it, so we trained linear probes using 3 subsets of these features and tested on the held out set.
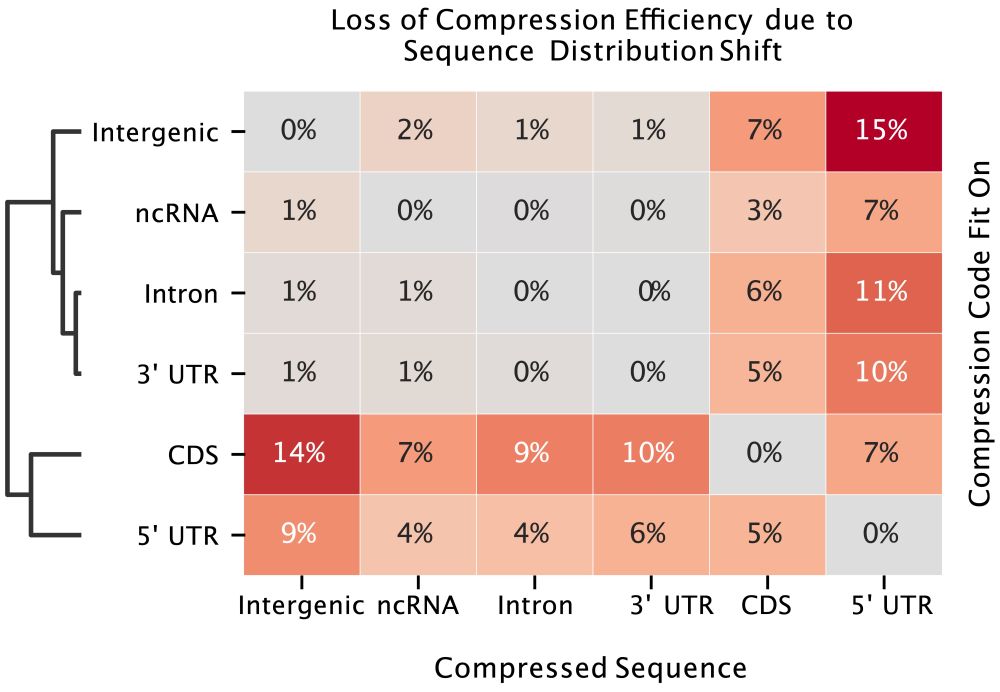
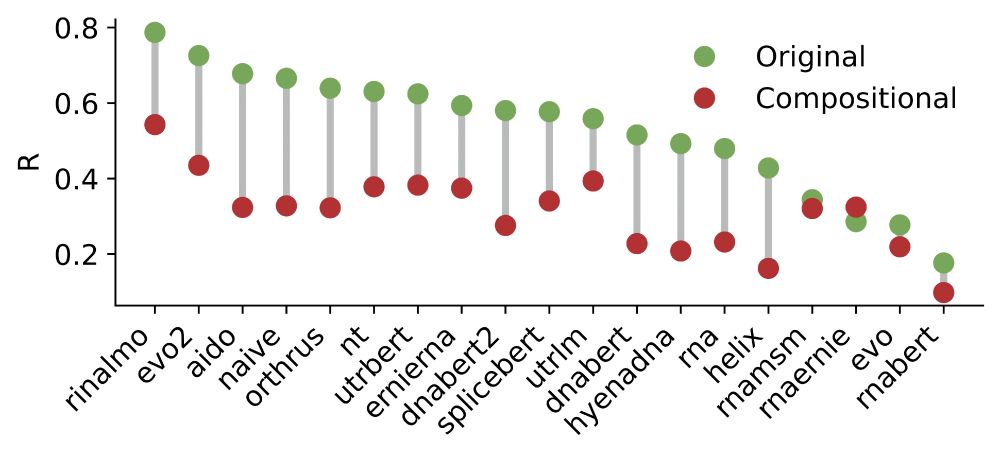
✅ Orthrus+MLM matches/beats SOTA on 6/10 datasets without increasing the training data or significantly increasing model parameters
📈 Pareto-dominates all models larger than 10 million parameters, including Evo2
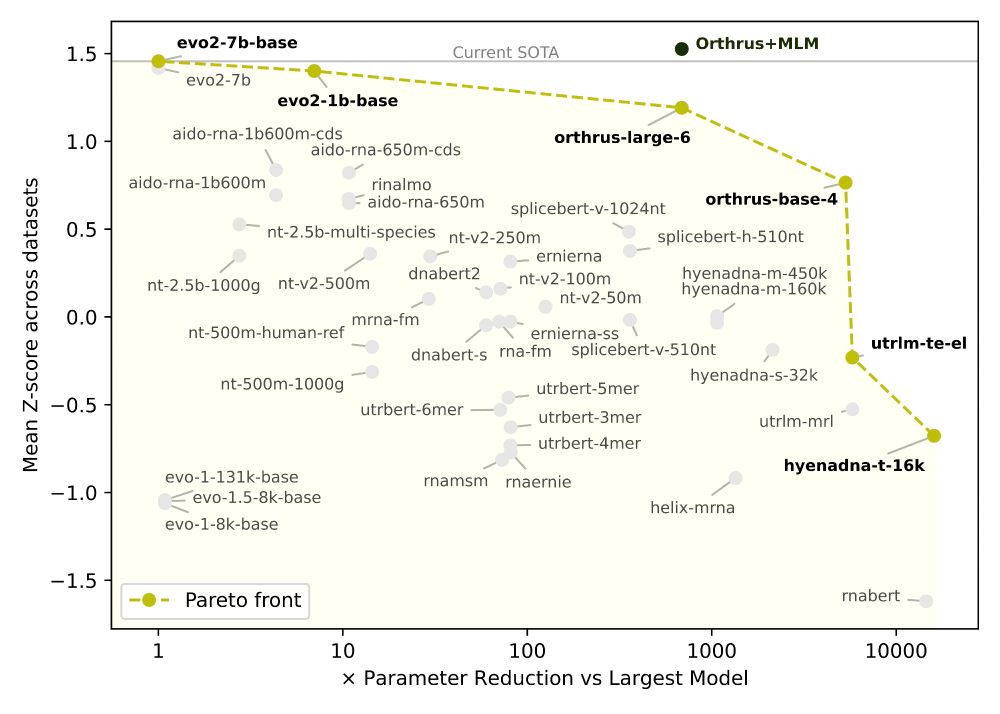
✅ Orthrus+MLM matches/beats SOTA on 6/10 datasets without increasing the training data or significantly increasing model parameters
📈 Pareto-dominates all models larger than 10 million parameters, including Evo2
Interestingly, we noticed that the 2nd best model, Orthrus, was not far off compared to Evo2 despite having only 10 million parameters
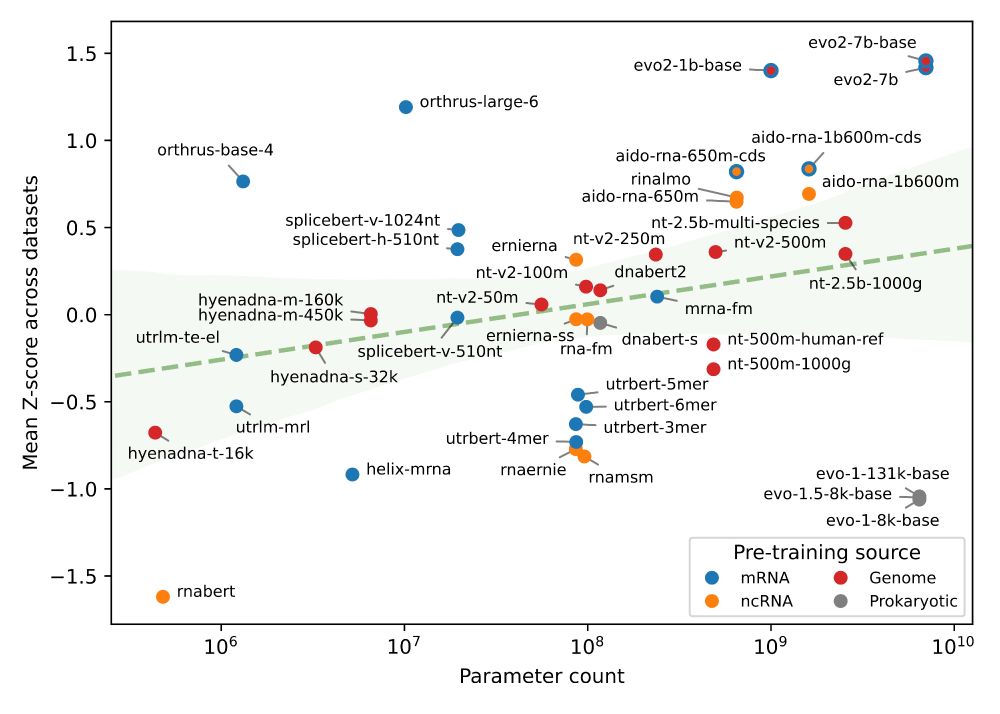
Interestingly, we noticed that the 2nd best model, Orthrus, was not far off compared to Evo2 despite having only 10 million parameters