
Steven Robbins
@stevenjrobbins.bsky.social
Do my science @ace_uq studying coral reef microbiomes. Data wrangler, meta-omics and long-read wonk, clean energy enthusiast, Saganist zealot, collector of weird zoology facts, other nonsense.
I also love stuff like this.

August 14, 2025 at 2:25 AM
I also love stuff like this.
This was a real prompt I gave chatgpt, with hilarious results.

August 13, 2025 at 10:41 AM
This was a real prompt I gave chatgpt, with hilarious results.
Yeah, if casting a wide net, Virsorter2 is probably the widest single-tool net, UNLESS you want to run PPR-Meta, which identifies a huge number of "viruses" unique to that tool, with no real indication they're erroneous as shown by plotting geNomad's marker genes in a PCA.

July 16, 2025 at 7:25 PM
Yeah, if casting a wide net, Virsorter2 is probably the widest single-tool net, UNLESS you want to run PPR-Meta, which identifies a huge number of "viruses" unique to that tool, with no real indication they're erroneous as shown by plotting geNomad's marker genes in a PCA.
But what we also noticed is that some core of viruses are predicted by multiple tools (rarely all), and TONS that are unique to each tool, especially PPR-Meta and VirSorter2. This venn diagram is from before we tweaked some settings, but shows the gist if you run things on default.

July 16, 2025 at 7:13 PM
But what we also noticed is that some core of viruses are predicted by multiple tools (rarely all), and TONS that are unique to each tool, especially PPR-Meta and VirSorter2. This venn diagram is from before we tweaked some settings, but shows the gist if you run things on default.
So we moved forward with our true Crassvirales contigs, made sure they had all the right Crassvirales core genes, and had a look at their taxonomy, incorporating several recently published Antarctic marine sequences from @goncalopiedade.bsky.social et al (2024).
pmc.ncbi.nlm.nih.gov/articles/PMC...
pmc.ncbi.nlm.nih.gov/articles/PMC...

July 16, 2025 at 6:22 PM
So we moved forward with our true Crassvirales contigs, made sure they had all the right Crassvirales core genes, and had a look at their taxonomy, incorporating several recently published Antarctic marine sequences from @goncalopiedade.bsky.social et al (2024).
pmc.ncbi.nlm.nih.gov/articles/PMC...
pmc.ncbi.nlm.nih.gov/articles/PMC...
We saw this same issue with ML-based "deep" classifiers for plasmids. DeepPlasmid, PlasClass, and Mobile-OG-db showed similar results when plotting Genomad's plasmid, chromosomal, viral markers. Contigs pred by these tools showed higher enrichment in chromosomal and viral markers than other tools.

June 10, 2025 at 11:35 AM
We saw this same issue with ML-based "deep" classifiers for plasmids. DeepPlasmid, PlasClass, and Mobile-OG-db showed similar results when plotting Genomad's plasmid, chromosomal, viral markers. Contigs pred by these tools showed higher enrichment in chromosomal and viral markers than other tools.
What's also interesting is that if you look at the same plot for Illumina-only metagenomes, this issue becomes much less pronounced, simply because the contigs are much shorter and do not often reach the range of erroneous assignment. So on short-read metagenomes, DeepVirFinder/CheckV may be fine.

June 10, 2025 at 11:19 AM
What's also interesting is that if you look at the same plot for Illumina-only metagenomes, this issue becomes much less pronounced, simply because the contigs are much shorter and do not often reach the range of erroneous assignment. So on short-read metagenomes, DeepVirFinder/CheckV may be fine.
Most meta-omic viral identification tools are tested on short-read metagenomes. We wanted to see if any looked wonky on long-read contigs. Most tools hold up, DeepVirFinder didn't. Plot shows the ratio of CheckV host to viral markers vs contig length for ONT assemblies, colored by CheckV quality.

June 10, 2025 at 9:39 AM
Most meta-omic viral identification tools are tested on short-read metagenomes. We wanted to see if any looked wonky on long-read contigs. Most tools hold up, DeepVirFinder didn't. Plot shows the ratio of CheckV host to viral markers vs contig length for ONT assemblies, colored by CheckV quality.
We noticed that another feature common to all our Ghost Taxa was low-GC (<40%). Graphing GC and Straininess together, we find a region of the graph at the intersection of high strain diversity and low-GC where Illumina short reads simply cannot go. And we see that our Ghost Taxa sit in that region.

May 25, 2025 at 10:38 AM
We noticed that another feature common to all our Ghost Taxa was low-GC (<40%). Graphing GC and Straininess together, we find a region of the graph at the intersection of high strain diversity and low-GC where Illumina short reads simply cannot go. And we see that our Ghost Taxa sit in that region.
Short-read metagenomic sequencing cannot recover genomes from many abundant marine prokaryotes due to high strain heterogeneity and platform-inherent GC bias (likely viruses, too), but Nanopore long reads can address this. A results thread on our recent preprint 🧵.

May 25, 2025 at 10:27 AM
Short-read metagenomic sequencing cannot recover genomes from many abundant marine prokaryotes due to high strain heterogeneity and platform-inherent GC bias (likely viruses, too), but Nanopore long reads can address this. A results thread on our recent preprint 🧵.
Feels like it belongs here

May 18, 2025 at 12:12 AM
Feels like it belongs here
For example, this is a bit old, but the general distribution still accurate. Lots of ONT reads still noisy.

April 9, 2025 at 2:19 AM
For example, this is a bit old, but the general distribution still accurate. Lots of ONT reads still noisy.
Hi all! Please check out @markoterzin.bsky.social's new paper in Microbiome investigating the relative reliability of taxonomy vs metagenomic gene predictions as bioindicators of 17 physicochemical variables (temp, nitrogen, etc) on the Great Barrier Reef.
Spoiler: genes > taxonomy.
Link below.
Spoiler: genes > taxonomy.
Link below.
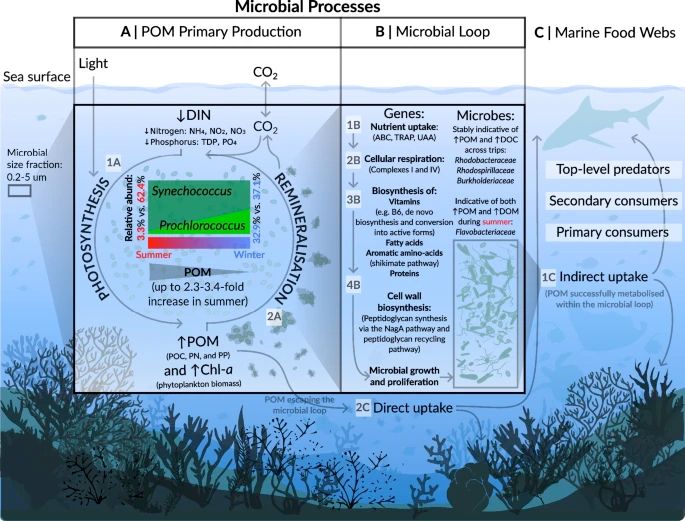
January 17, 2025 at 9:00 PM
Hi all! Please check out @markoterzin.bsky.social's new paper in Microbiome investigating the relative reliability of taxonomy vs metagenomic gene predictions as bioindicators of 17 physicochemical variables (temp, nitrogen, etc) on the Great Barrier Reef.
Spoiler: genes > taxonomy.
Link below.
Spoiler: genes > taxonomy.
Link below.
My Halloween costume this year. Deep sea creatures are kind of like aliens?

November 10, 2024 at 10:34 PM
My Halloween costume this year. Deep sea creatures are kind of like aliens?

