
Sternberg Lab
@sternberglab.bsky.social
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
10/10 Many thanks to all co-authors for their collective efforts in bringing this story to fruition! And a special thanks to @shsternberg.bsky.social for continually fostering a spirit of curiosity, creativity, and rigor in all aspects of our work.
October 17, 2025 at 5:22 PM
10/10 Many thanks to all co-authors for their collective efforts in bringing this story to fruition! And a special thanks to @shsternberg.bsky.social for continually fostering a spirit of curiosity, creativity, and rigor in all aspects of our work.
9/10 Collectively, our findings suggest that telomerase-like activity emerged in an ancient bacterial ancestor, and was co-opted in early organisms with linear genomes to set the stage for the evolution of modern eukaryotes.
October 17, 2025 at 5:22 PM
9/10 Collectively, our findings suggest that telomerase-like activity emerged in an ancient bacterial ancestor, and was co-opted in early organisms with linear genomes to set the stage for the evolution of modern eukaryotes.
8/10 Furthermore, when equipped with the template sequence from the telomerase RNA, DRT10 readily synthesized telomeric DNA repeats.

October 17, 2025 at 5:22 PM
8/10 Furthermore, when equipped with the template sequence from the telomerase RNA, DRT10 readily synthesized telomeric DNA repeats.
7/10 We teamed up with RT aficionado @pentamorfico.bsky.social from the Rafa Pinilla-Redondo lab to build a new phylogenetic tree of RTs across all domains of life.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.

October 17, 2025 at 5:22 PM
7/10 We teamed up with RT aficionado @pentamorfico.bsky.social from the Rafa Pinilla-Redondo lab to build a new phylogenetic tree of RTs across all domains of life.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
6/10 We found that DRT10 synthesizes tandem-repeat cDNAs through a mechanism strikingly reminiscent of DNA repeat addition by telomerase. But does this similarity represent convergent evolution or shared ancestry?

October 17, 2025 at 5:22 PM
6/10 We found that DRT10 synthesizes tandem-repeat cDNAs through a mechanism strikingly reminiscent of DNA repeat addition by telomerase. But does this similarity represent convergent evolution or shared ancestry?
5/10 Following our recent work on the DRT2 and DRT9 antiviral immune systems in bacteria (linked below), which revealed intricate mechanisms of RNA-templated repetitive DNA synthesis, we began studying a new system, DRT10.
DRT2: science.org/doi/10.1126/...
DRT9: nature.com/articles/s41...
DRT2: science.org/doi/10.1126/...
DRT9: nature.com/articles/s41...

De novo gene synthesis by an antiviral reverse transcriptase
Defense-associated reverse transcriptase (DRT) systems perform DNA synthesis to protect bacteria against viral infection, but the identities and functions of their DNA products remain largely unknown....
science.org
October 17, 2025 at 5:22 PM
5/10 Following our recent work on the DRT2 and DRT9 antiviral immune systems in bacteria (linked below), which revealed intricate mechanisms of RNA-templated repetitive DNA synthesis, we began studying a new system, DRT10.
DRT2: science.org/doi/10.1126/...
DRT9: nature.com/articles/s41...
DRT2: science.org/doi/10.1126/...
DRT9: nature.com/articles/s41...
4/10 Telomerase is found in nearly all eukaryotes and acts as a critical safeguard against genome instability from progressive DNA loss. Its aberrant activation is also key to the growth of cancer cells.
And yet, the evolutionary origin of telomerase has long remained unresolved.
And yet, the evolutionary origin of telomerase has long remained unresolved.
October 17, 2025 at 5:22 PM
4/10 Telomerase is found in nearly all eukaryotes and acts as a critical safeguard against genome instability from progressive DNA loss. Its aberrant activation is also key to the growth of cancer cells.
And yet, the evolutionary origin of telomerase has long remained unresolved.
And yet, the evolutionary origin of telomerase has long remained unresolved.
3/10 Forty years ago, Carol Greider and Elizabeth Blackburn discovered an enzyme that solves this problem. Telomerase, which comprises a reverse transcriptase (TERT) and RNA (TR), directly extends chromosome ends by adding DNA repeats templated by the TR.

October 17, 2025 at 5:22 PM
3/10 Forty years ago, Carol Greider and Elizabeth Blackburn discovered an enzyme that solves this problem. Telomerase, which comprises a reverse transcriptase (TERT) and RNA (TR), directly extends chromosome ends by adding DNA repeats templated by the TR.
2/10 Linear chromosomes shorten with each round of cell division. Famously known as the “end-replication problem,” this phenomenon eventually leads to cellular senescence and is one of the hallmarks of aging.
October 17, 2025 at 5:22 PM
2/10 Linear chromosomes shorten with each round of cell division. Famously known as the “end-replication problem,” this phenomenon eventually leads to cellular senescence and is one of the hallmarks of aging.
16/16 This work was a big team effort and fun collaboration! Thanks to everyone: Matt, Egill, Tanner, Jing, Zaofeng, Americo, Florian, Hamna, Ryan, Nick, Israel and our stellar mentor Sam for making this project possible!
July 24, 2025 at 7:58 PM
16/16 This work was a big team effort and fun collaboration! Thanks to everyone: Matt, Egill, Tanner, Jing, Zaofeng, Americo, Florian, Hamna, Ryan, Nick, Israel and our stellar mentor Sam for making this project possible!
15/16 In summary, we shed light on the biological role of a naturally occurring version of CRISPR interference (CRISPRi), that emerged in gut-associated bacteria to benefit multiple aspects of their lifestyle.
July 24, 2025 at 7:58 PM
15/16 In summary, we shed light on the biological role of a naturally occurring version of CRISPR interference (CRISPRi), that emerged in gut-associated bacteria to benefit multiple aspects of their lifestyle.
14/16 The repeated exaptation of TnpB to regulate flagellin suggests strong evolutionary pressure favoring RNA-guided control of flagellin expression, particularly in gut-associated bacterial species.
July 24, 2025 at 7:58 PM
14/16 The repeated exaptation of TnpB to regulate flagellin suggests strong evolutionary pressure favoring RNA-guided control of flagellin expression, particularly in gut-associated bacterial species.
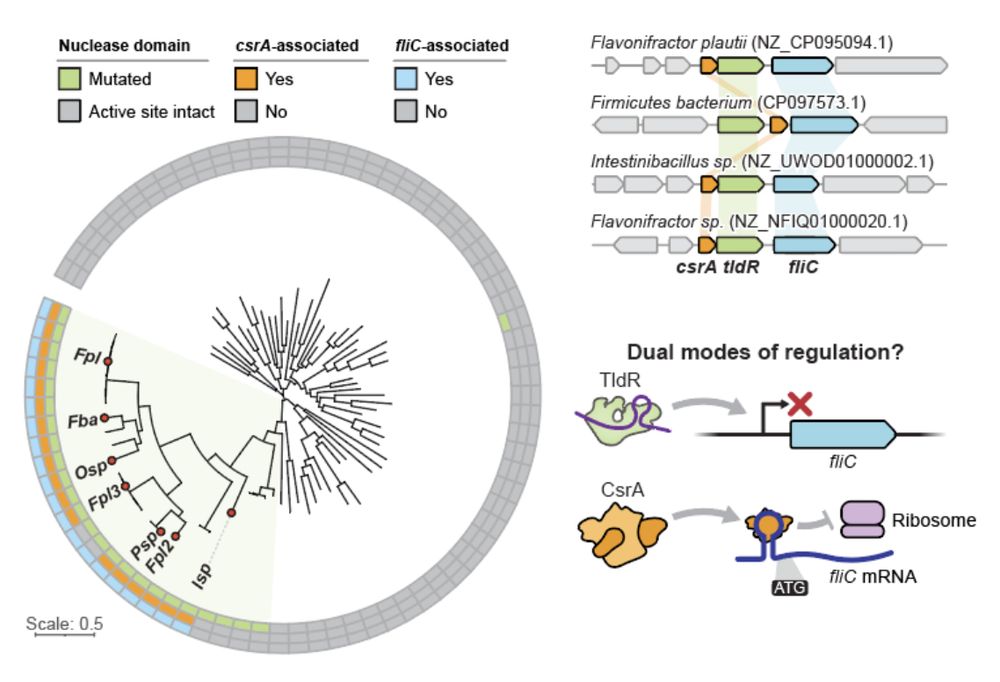
13/16 In other key members of the human gut microbiome, flagellin-regulating TldRs can be found additionally associated with the translational repressor CsrA. The coordinated action of TldR and CsrA suggests a dual mode of both transcriptional and post-transcriptional gene regulation.
July 24, 2025 at 7:58 PM
13/16 In other key members of the human gut microbiome, flagellin-regulating TldRs can be found additionally associated with the translational repressor CsrA. The coordinated action of TldR and CsrA suggests a dual mode of both transcriptional and post-transcriptional gene regulation.
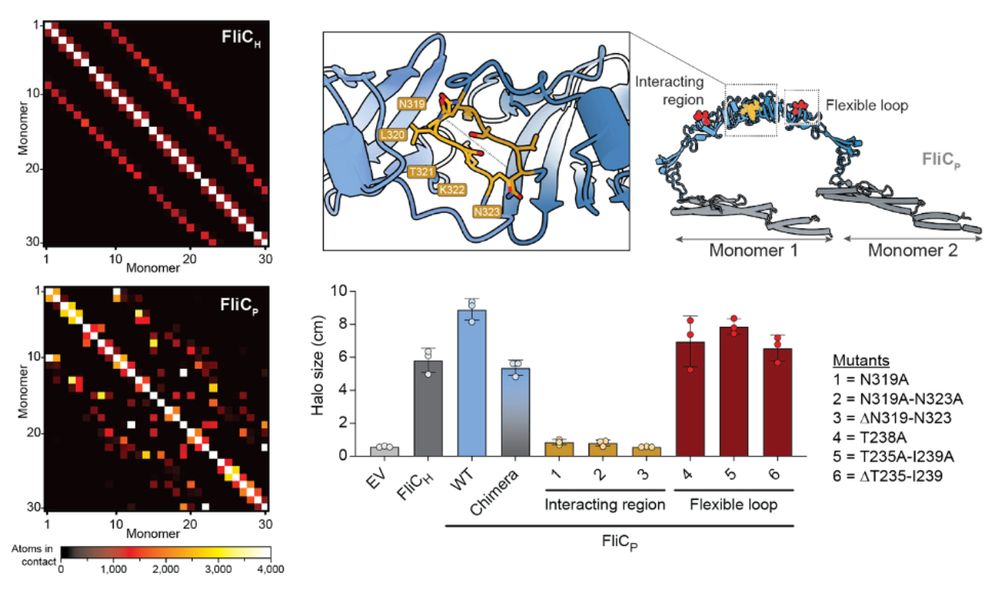
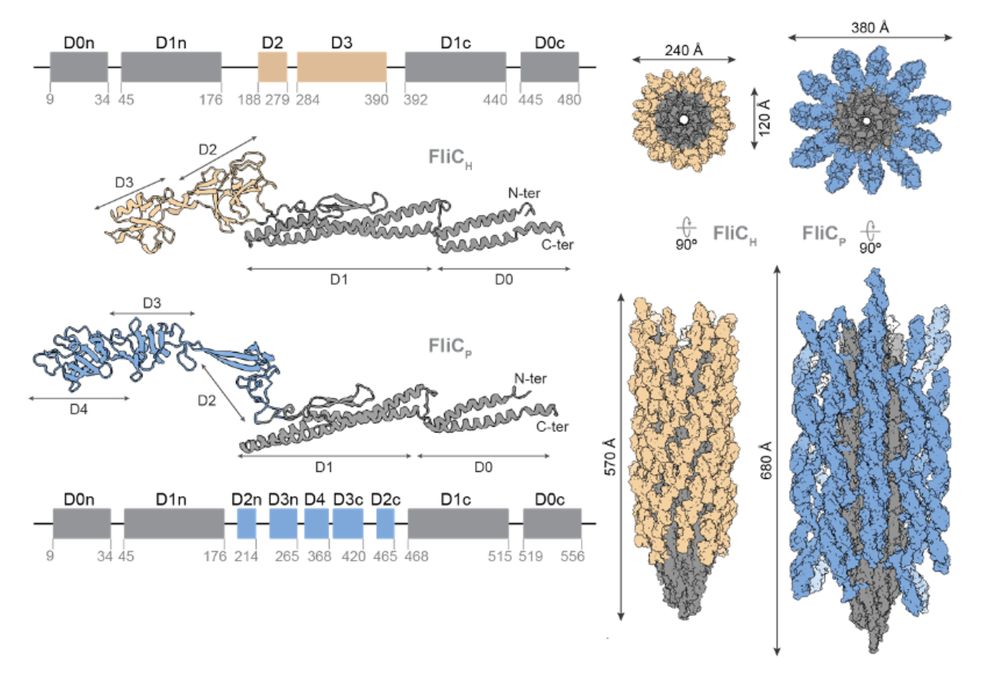
12/16 The extra domain stabilizes the filament by increasing inter-subunit contacts. Structure-guided mutagenesis confirmed that this domain is essential to improve motility of the lysogen. We believe it could similarly explain the decreased activation of TLR5.
July 24, 2025 at 7:58 PM
12/16 The extra domain stabilizes the filament by increasing inter-subunit contacts. Structure-guided mutagenesis confirmed that this domain is essential to improve motility of the lysogen. We believe it could similarly explain the decreased activation of TLR5.
11/16 We found that both flagellar filaments have a drastically different structure. The prophage flagellar filament is significantly thicker and presents a surprising mesh-like structure. This is due to the presence of an extra domain in the prophage flagellin.
July 24, 2025 at 7:58 PM
11/16 We found that both flagellar filaments have a drastically different structure. The prophage flagellar filament is significantly thicker and presents a surprising mesh-like structure. This is due to the presence of an extra domain in the prophage flagellin.
10/16 To try to link these phenotypic observations, we teamed up once again with @piratefernandez.bsky.social and decided to inspect the structures of the prophage flagellin in comparison to their host counterpart.
July 24, 2025 at 7:58 PM
10/16 To try to link these phenotypic observations, we teamed up once again with @piratefernandez.bsky.social and decided to inspect the structures of the prophage flagellin in comparison to their host counterpart.
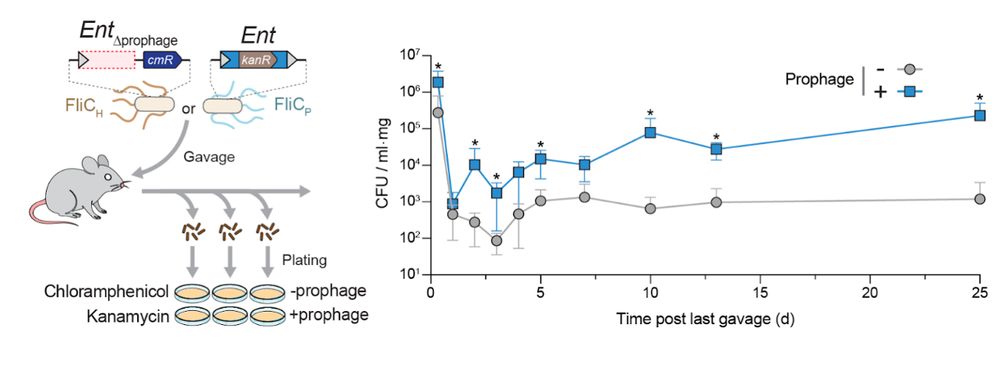
9/16 The phenotypic effects of lysogenization result in a consequential ecological outcome: enhanced colonization of the murine gut, illustrating the advantage of flagellar remodeling in a host-associated environment.
July 24, 2025 at 7:58 PM
9/16 The phenotypic effects of lysogenization result in a consequential ecological outcome: enhanced colonization of the murine gut, illustrating the advantage of flagellar remodeling in a host-associated environment.
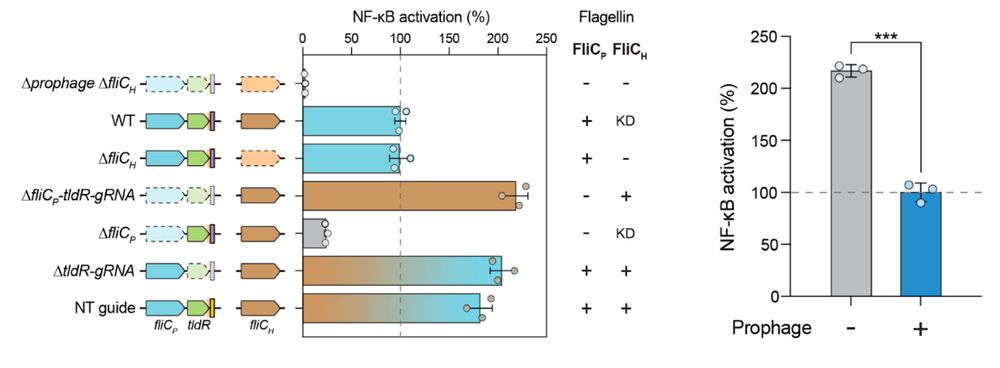
8/16 We also find that lysogenic conversion by FRφ reduces activation of TLR5, a receptor that detects flagellin to initiate the immune response, suggesting that lysogenization may facilitate immune evasion.
July 24, 2025 at 7:58 PM
8/16 We also find that lysogenic conversion by FRφ reduces activation of TLR5, a receptor that detects flagellin to initiate the immune response, suggesting that lysogenization may facilitate immune evasion.
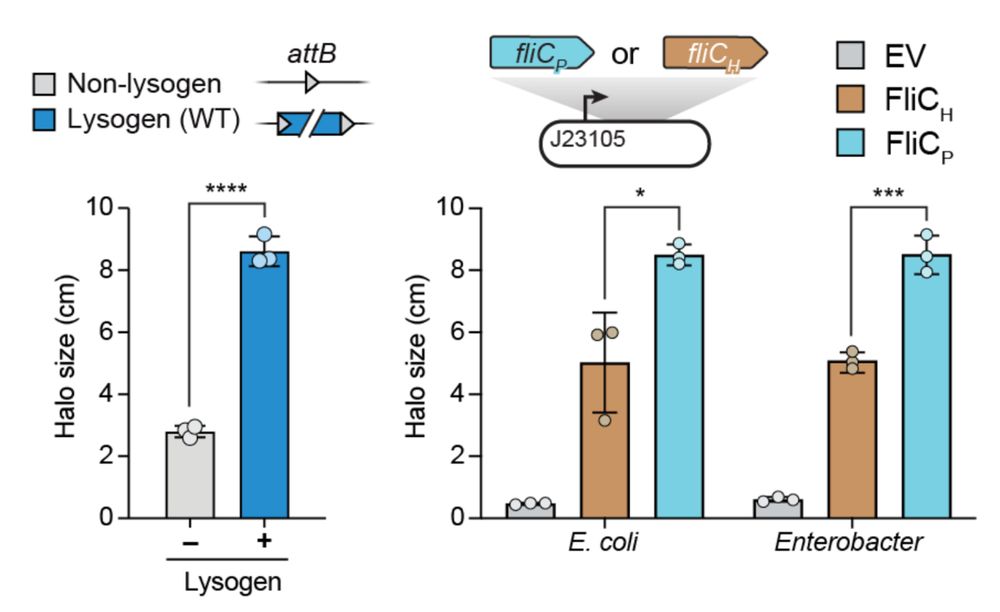
7/16 We find that lysogenic conversion by FRφ leads to increased motility of the host. Essentially, Enterobacter (and even E. coli actually) swims faster when expressing FliCp instead of FliCh.
July 24, 2025 at 7:58 PM
7/16 We find that lysogenic conversion by FRφ leads to increased motility of the host. Essentially, Enterobacter (and even E. coli actually) swims faster when expressing FliCp instead of FliCh.
6/16 To tackle this question we focused on a clinical isolate of Enterobacter, that natively contains what we now call a Flagellin Remodeling prophage (FRφ), and started our detective work to uncover the potential advantage(s) conferred by the acquisition of this prophage.
July 24, 2025 at 7:58 PM
6/16 To tackle this question we focused on a clinical isolate of Enterobacter, that natively contains what we now call a Flagellin Remodeling prophage (FRφ), and started our detective work to uncover the potential advantage(s) conferred by the acquisition of this prophage.
5/16 Prophages sometimes provide new traits to their bacterial host, that is called “lysogenic conversion”. Now, why would a phage manipulate the flagellar apparatus of their host? That question is the focus of our new study.
July 24, 2025 at 7:58 PM
5/16 Prophages sometimes provide new traits to their bacterial host, that is called “lysogenic conversion”. Now, why would a phage manipulate the flagellar apparatus of their host? That question is the focus of our new study.
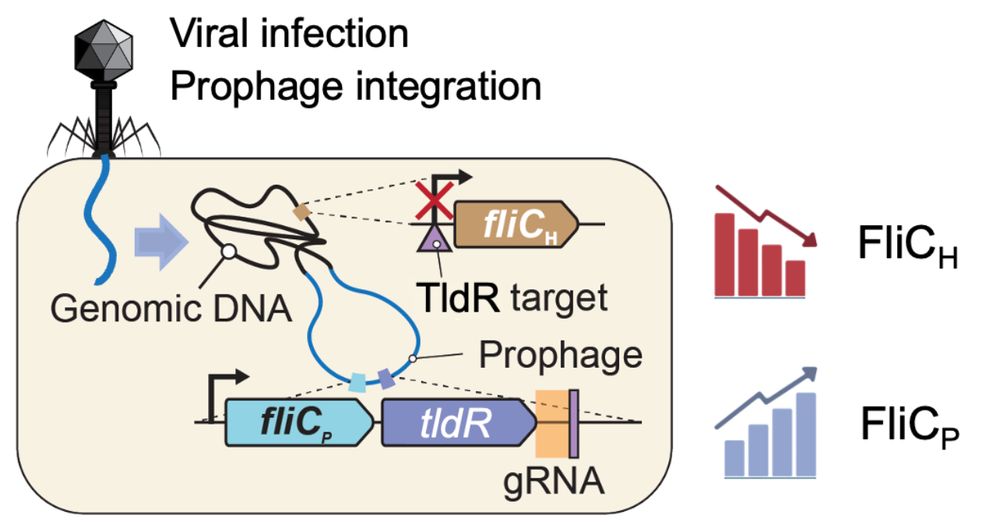
4/16 Strikingly, FliC-associated TldRs are found in prophages and target the promoter of host-encoded FliC. Basically, TldR silences the host FliC (FliCh) and the prophage copy (FliCp) takes over, effectively leading to a remodeling of the host flagellar apparatus.
July 24, 2025 at 7:58 PM
4/16 Strikingly, FliC-associated TldRs are found in prophages and target the promoter of host-encoded FliC. Basically, TldR silences the host FliC (FliCh) and the prophage copy (FliCp) takes over, effectively leading to a remodeling of the host flagellar apparatus.
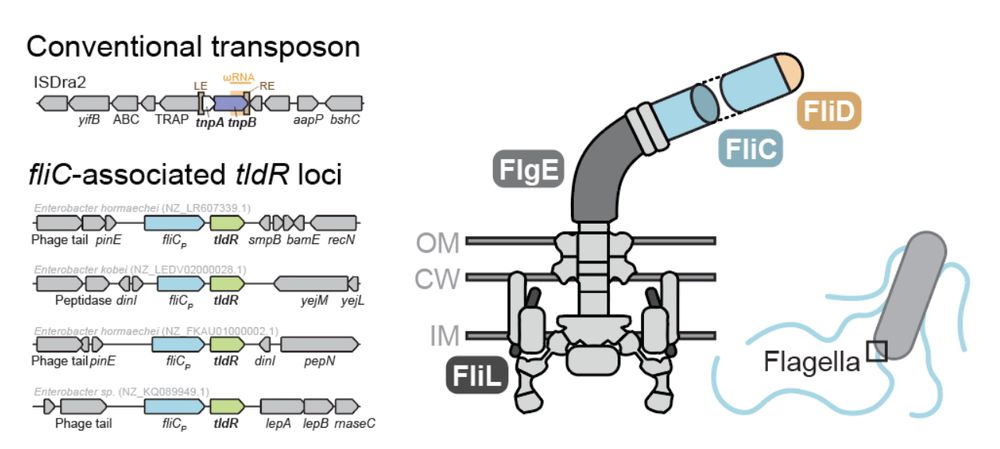
3/16 Unlike TnpB, which can be found in transposons and in association with transposases, TldR is often found in association with FliC (flagellin), the main subunit of flagellar filaments. Flagella are large appendages used by bacteria for motility.
July 24, 2025 at 7:58 PM
3/16 Unlike TnpB, which can be found in transposons and in association with transposases, TldR is often found in association with FliC (flagellin), the main subunit of flagellar filaments. Flagella are large appendages used by bacteria for motility.

