
Sternberg Lab
@sternberglab.bsky.social
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
8/10 Furthermore, when equipped with the template sequence from the telomerase RNA, DRT10 readily synthesized telomeric DNA repeats.

October 17, 2025 at 5:22 PM
8/10 Furthermore, when equipped with the template sequence from the telomerase RNA, DRT10 readily synthesized telomeric DNA repeats.
7/10 We teamed up with RT aficionado @pentamorfico.bsky.social from the Rafa Pinilla-Redondo lab to build a new phylogenetic tree of RTs across all domains of life.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.

October 17, 2025 at 5:22 PM
7/10 We teamed up with RT aficionado @pentamorfico.bsky.social from the Rafa Pinilla-Redondo lab to build a new phylogenetic tree of RTs across all domains of life.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
6/10 We found that DRT10 synthesizes tandem-repeat cDNAs through a mechanism strikingly reminiscent of DNA repeat addition by telomerase. But does this similarity represent convergent evolution or shared ancestry?

October 17, 2025 at 5:22 PM
6/10 We found that DRT10 synthesizes tandem-repeat cDNAs through a mechanism strikingly reminiscent of DNA repeat addition by telomerase. But does this similarity represent convergent evolution or shared ancestry?
3/10 Forty years ago, Carol Greider and Elizabeth Blackburn discovered an enzyme that solves this problem. Telomerase, which comprises a reverse transcriptase (TERT) and RNA (TR), directly extends chromosome ends by adding DNA repeats templated by the TR.

October 17, 2025 at 5:22 PM
3/10 Forty years ago, Carol Greider and Elizabeth Blackburn discovered an enzyme that solves this problem. Telomerase, which comprises a reverse transcriptase (TERT) and RNA (TR), directly extends chromosome ends by adding DNA repeats templated by the TR.
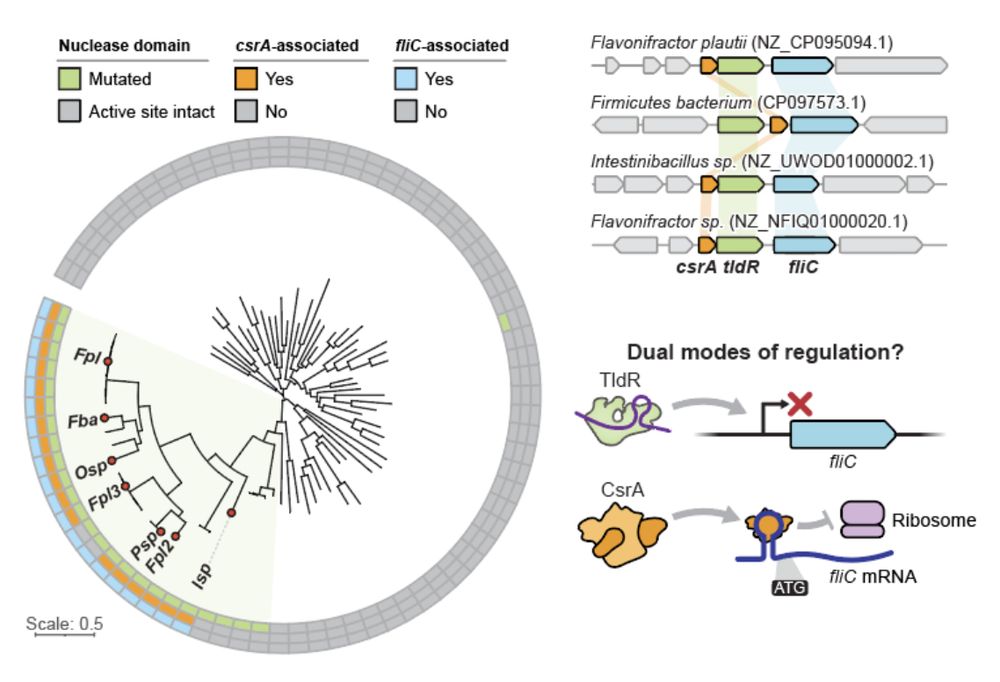
13/16 In other key members of the human gut microbiome, flagellin-regulating TldRs can be found additionally associated with the translational repressor CsrA. The coordinated action of TldR and CsrA suggests a dual mode of both transcriptional and post-transcriptional gene regulation.
July 24, 2025 at 7:58 PM
13/16 In other key members of the human gut microbiome, flagellin-regulating TldRs can be found additionally associated with the translational repressor CsrA. The coordinated action of TldR and CsrA suggests a dual mode of both transcriptional and post-transcriptional gene regulation.
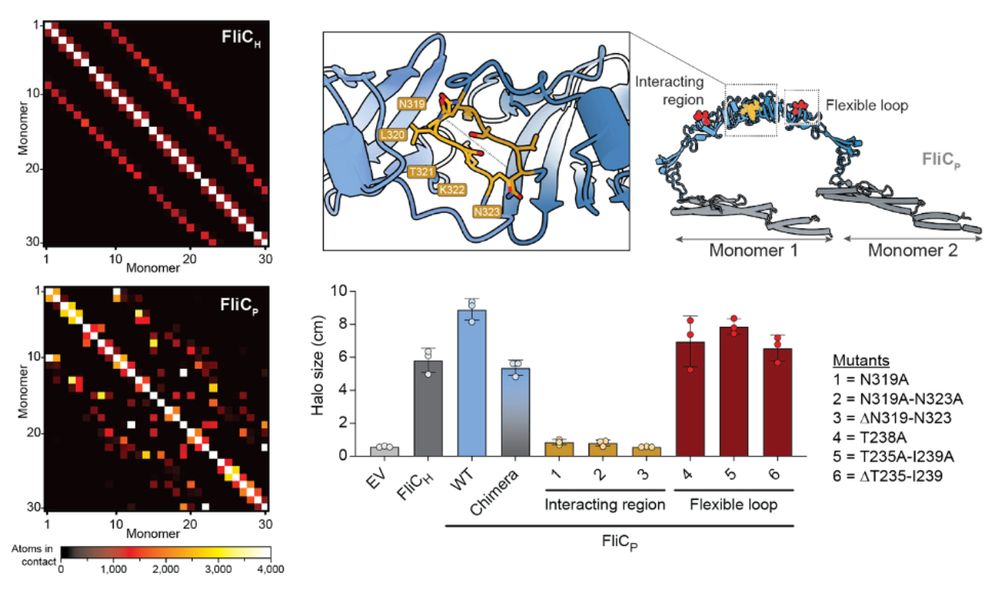
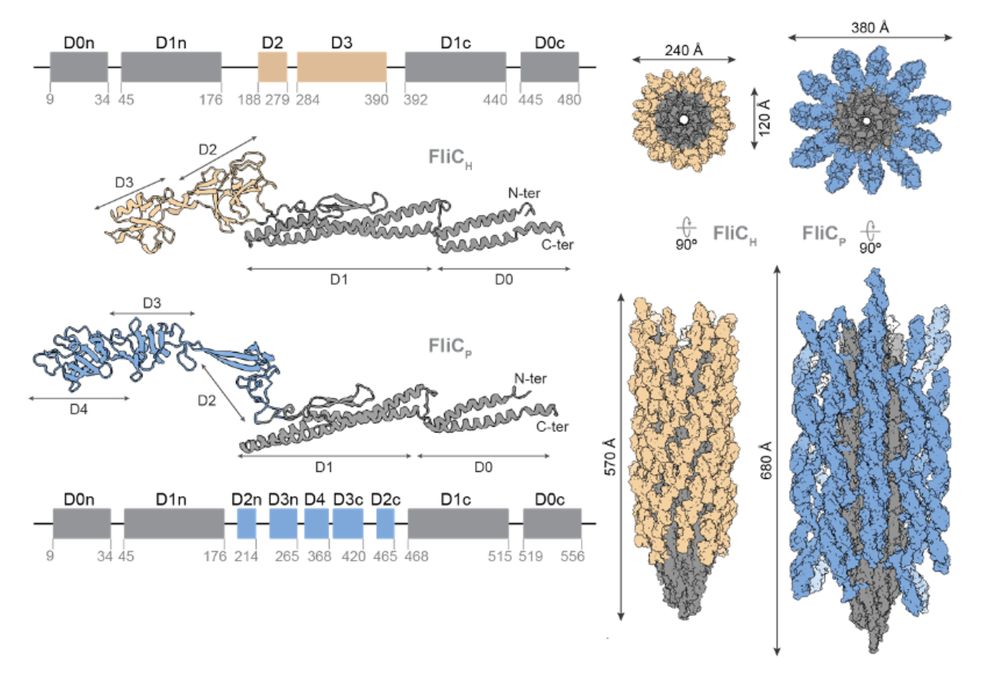
12/16 The extra domain stabilizes the filament by increasing inter-subunit contacts. Structure-guided mutagenesis confirmed that this domain is essential to improve motility of the lysogen. We believe it could similarly explain the decreased activation of TLR5.
July 24, 2025 at 7:58 PM
12/16 The extra domain stabilizes the filament by increasing inter-subunit contacts. Structure-guided mutagenesis confirmed that this domain is essential to improve motility of the lysogen. We believe it could similarly explain the decreased activation of TLR5.
11/16 We found that both flagellar filaments have a drastically different structure. The prophage flagellar filament is significantly thicker and presents a surprising mesh-like structure. This is due to the presence of an extra domain in the prophage flagellin.
July 24, 2025 at 7:58 PM
11/16 We found that both flagellar filaments have a drastically different structure. The prophage flagellar filament is significantly thicker and presents a surprising mesh-like structure. This is due to the presence of an extra domain in the prophage flagellin.
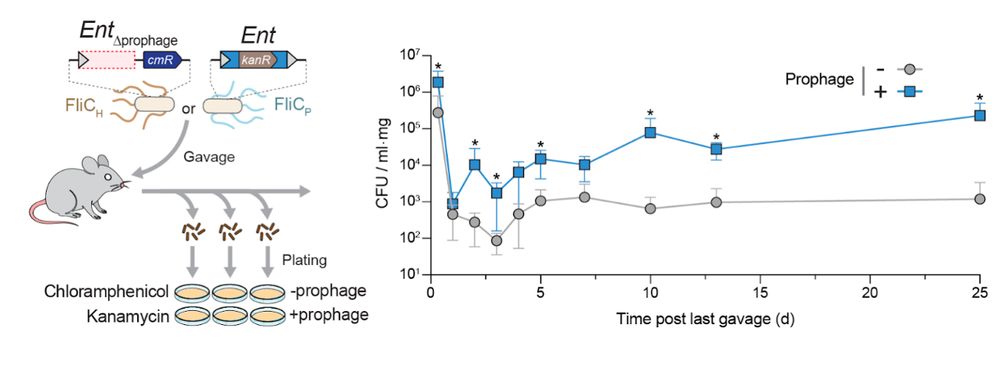
9/16 The phenotypic effects of lysogenization result in a consequential ecological outcome: enhanced colonization of the murine gut, illustrating the advantage of flagellar remodeling in a host-associated environment.
July 24, 2025 at 7:58 PM
9/16 The phenotypic effects of lysogenization result in a consequential ecological outcome: enhanced colonization of the murine gut, illustrating the advantage of flagellar remodeling in a host-associated environment.
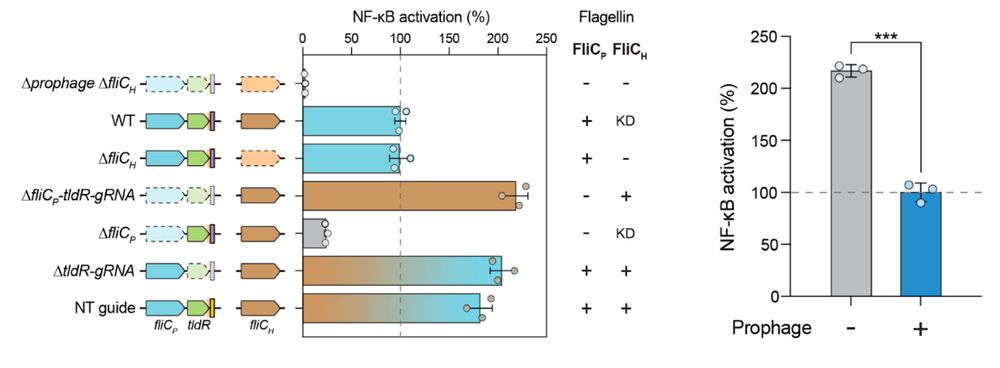
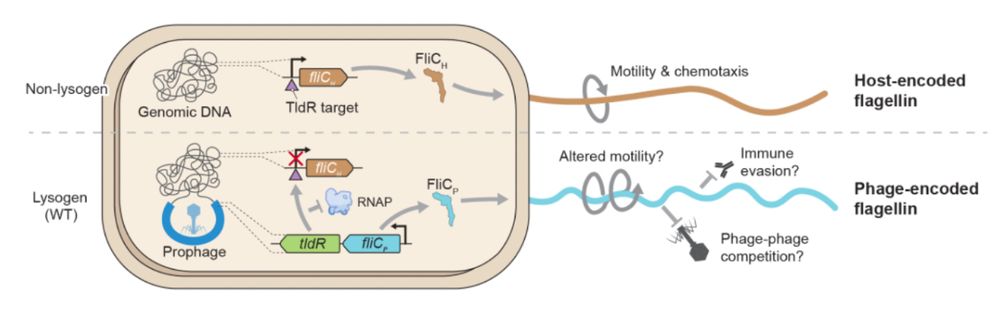
8/16 We also find that lysogenic conversion by FRφ reduces activation of TLR5, a receptor that detects flagellin to initiate the immune response, suggesting that lysogenization may facilitate immune evasion.
July 24, 2025 at 7:58 PM
8/16 We also find that lysogenic conversion by FRφ reduces activation of TLR5, a receptor that detects flagellin to initiate the immune response, suggesting that lysogenization may facilitate immune evasion.
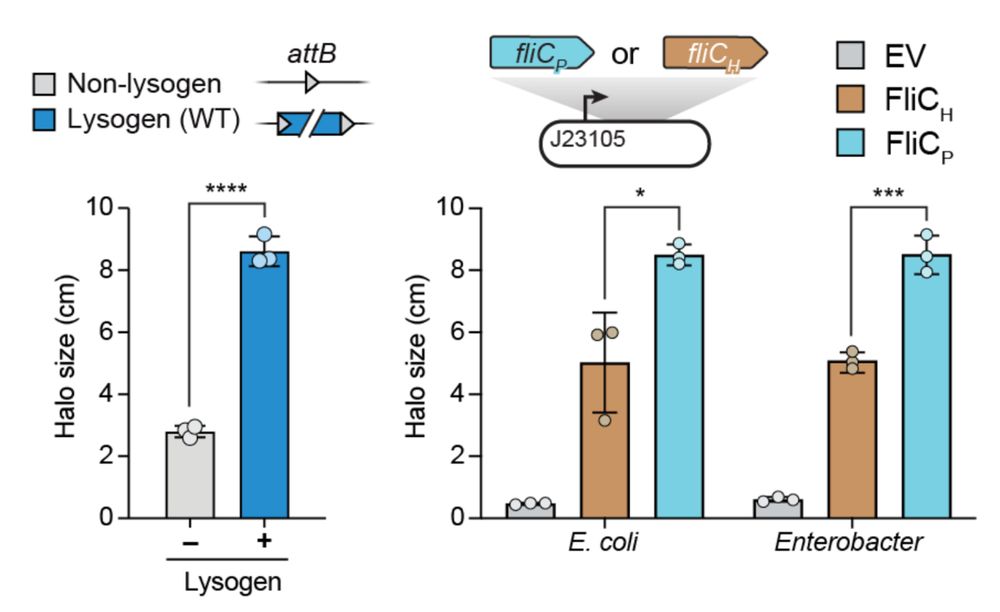
7/16 We find that lysogenic conversion by FRφ leads to increased motility of the host. Essentially, Enterobacter (and even E. coli actually) swims faster when expressing FliCp instead of FliCh.
July 24, 2025 at 7:58 PM
7/16 We find that lysogenic conversion by FRφ leads to increased motility of the host. Essentially, Enterobacter (and even E. coli actually) swims faster when expressing FliCp instead of FliCh.
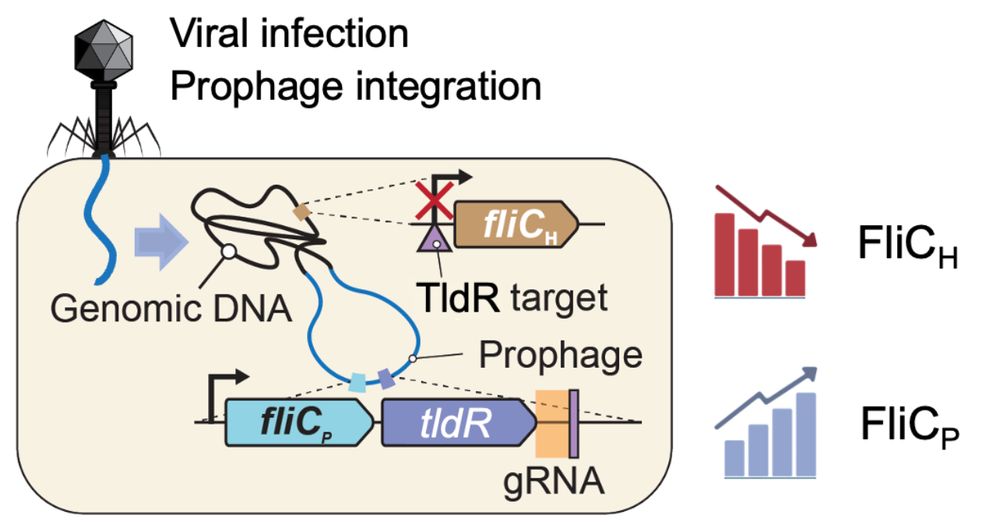
4/16 Strikingly, FliC-associated TldRs are found in prophages and target the promoter of host-encoded FliC. Basically, TldR silences the host FliC (FliCh) and the prophage copy (FliCp) takes over, effectively leading to a remodeling of the host flagellar apparatus.
July 24, 2025 at 7:58 PM
4/16 Strikingly, FliC-associated TldRs are found in prophages and target the promoter of host-encoded FliC. Basically, TldR silences the host FliC (FliCh) and the prophage copy (FliCp) takes over, effectively leading to a remodeling of the host flagellar apparatus.
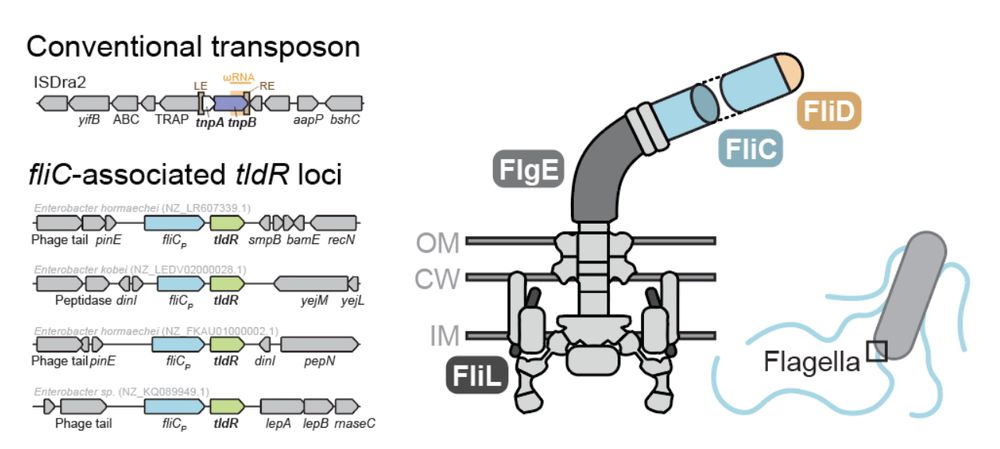
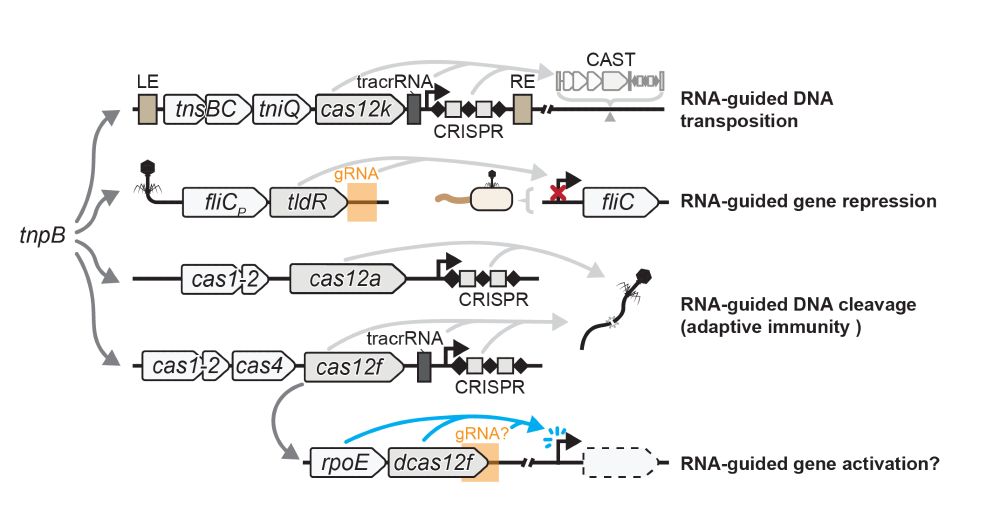
3/16 Unlike TnpB, which can be found in transposons and in association with transposases, TldR is often found in association with FliC (flagellin), the main subunit of flagellar filaments. Flagella are large appendages used by bacteria for motility.
July 24, 2025 at 7:58 PM
3/16 Unlike TnpB, which can be found in transposons and in association with transposases, TldR is often found in association with FliC (flagellin), the main subunit of flagellar filaments. Flagella are large appendages used by bacteria for motility.
2/16 Last year we reported the discovery of RNA-guided transcriptional repressors, derived from the Cas12 ancestor TnpB. We called it TldR for “TnpB-like nuclease dead repressors”. Yet, we did not address their actual biological role.
More details here: tinyurl.com/2camf88e
More details here: tinyurl.com/2camf88e
July 24, 2025 at 7:58 PM
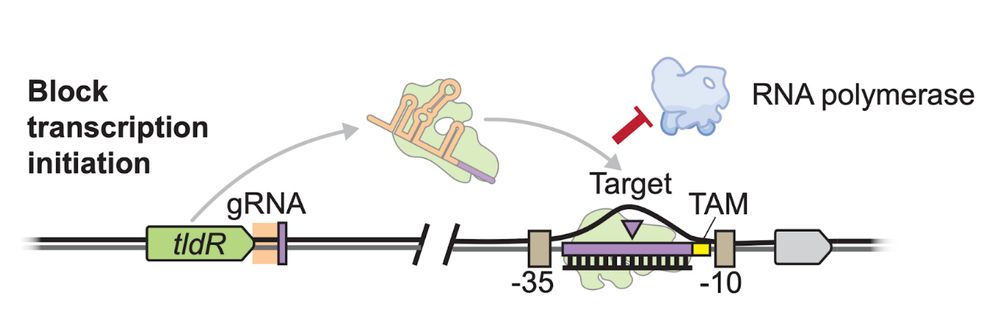
2/16 Last year we reported the discovery of RNA-guided transcriptional repressors, derived from the Cas12 ancestor TnpB. We called it TldR for “TnpB-like nuclease dead repressors”. Yet, we did not address their actual biological role.
More details here: tinyurl.com/2camf88e
More details here: tinyurl.com/2camf88e
1/16 New pre-print from the Sternberg Lab!
We uncover how temperate phages can use RNA-guided transcription factors to remodel the flagellar composition of their bacterial host and enhance their fitness.
Find the preprint and full story here: tinyurl.com/mshwjd77
We uncover how temperate phages can use RNA-guided transcription factors to remodel the flagellar composition of their bacterial host and enhance their fitness.
Find the preprint and full story here: tinyurl.com/mshwjd77
July 24, 2025 at 7:58 PM
1/16 New pre-print from the Sternberg Lab!
We uncover how temperate phages can use RNA-guided transcription factors to remodel the flagellar composition of their bacterial host and enhance their fitness.
Find the preprint and full story here: tinyurl.com/mshwjd77
We uncover how temperate phages can use RNA-guided transcription factors to remodel the flagellar composition of their bacterial host and enhance their fitness.
Find the preprint and full story here: tinyurl.com/mshwjd77
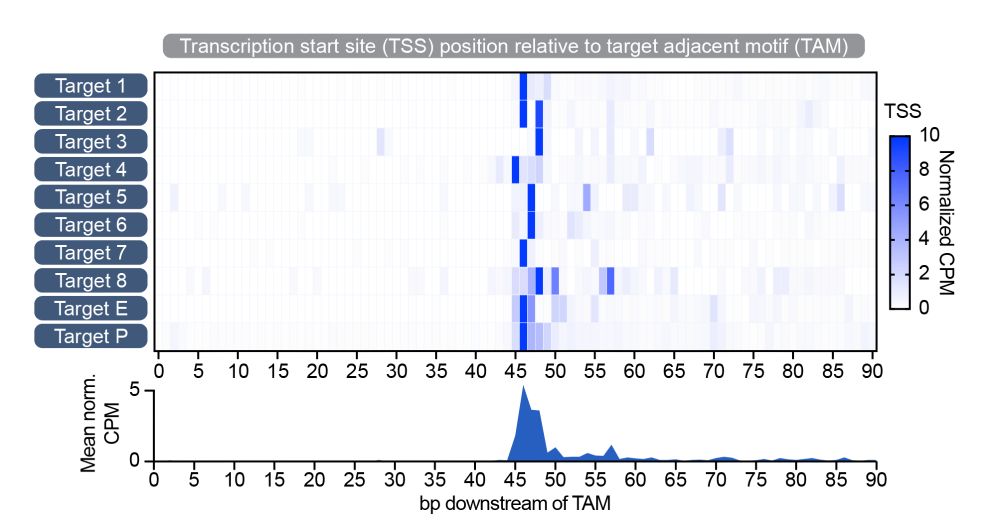
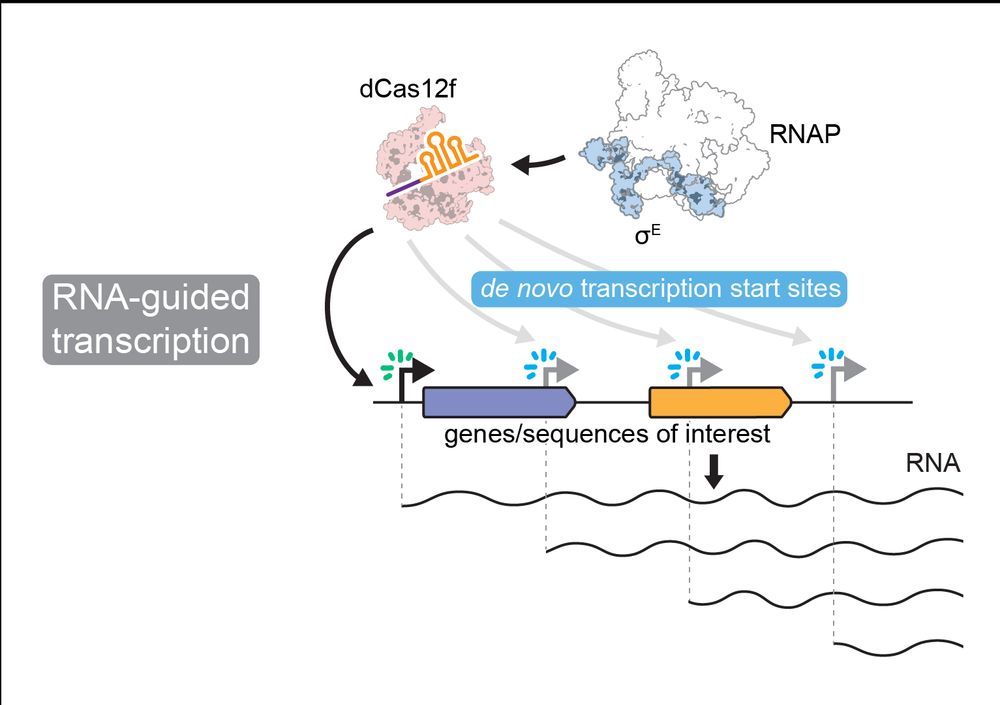
7/10 Transcription initiates in a tight ~45-48 bp window downstream of the PAM/TAM flanking the target site. All sequence specificity is imparted by Cas12f. The associated Sigma factor is recruited without imposing any of the typical -35 and -10 promoter DNA sequence requirements.
June 11, 2025 at 4:03 PM
7/10 Transcription initiates in a tight ~45-48 bp window downstream of the PAM/TAM flanking the target site. All sequence specificity is imparted by Cas12f. The associated Sigma factor is recruited without imposing any of the typical -35 and -10 promoter DNA sequence requirements.
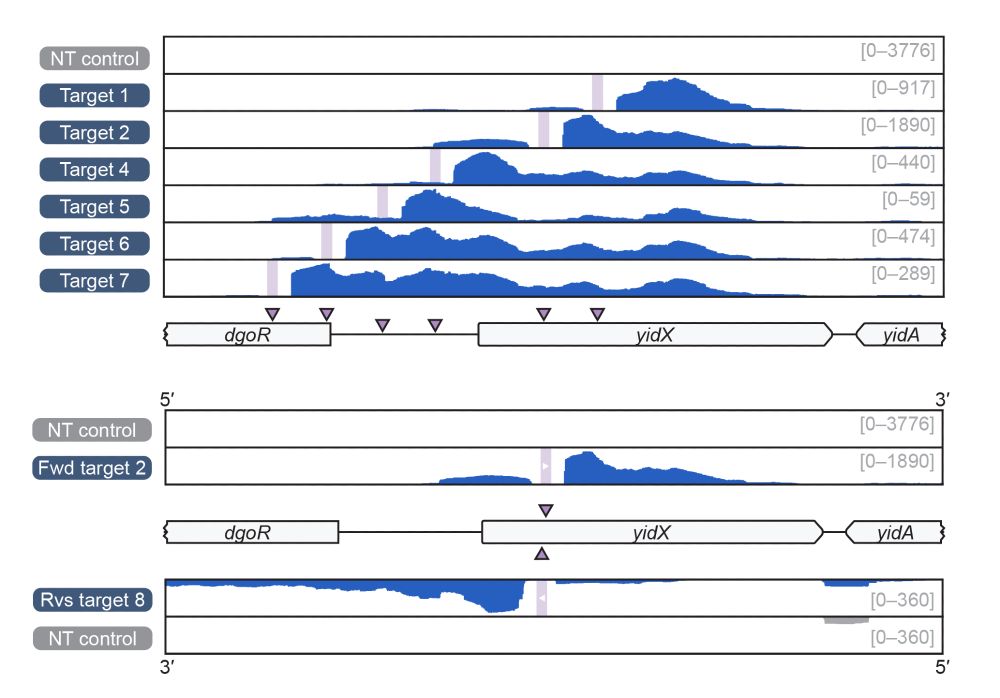
6/10 dCas12f-Sigma factors are able to recruit their native RNA polymerase (RNAP) to drive RNA-guided transcription at programmable sites. Remarkably, we were able to generate RNA (see RNA-seq) without relying on promoters in intergenic sequences, even within genes, and on either DNA strand.
June 11, 2025 at 4:03 PM
6/10 dCas12f-Sigma factors are able to recruit their native RNA polymerase (RNAP) to drive RNA-guided transcription at programmable sites. Remarkably, we were able to generate RNA (see RNA-seq) without relying on promoters in intergenic sequences, even within genes, and on either DNA strand.
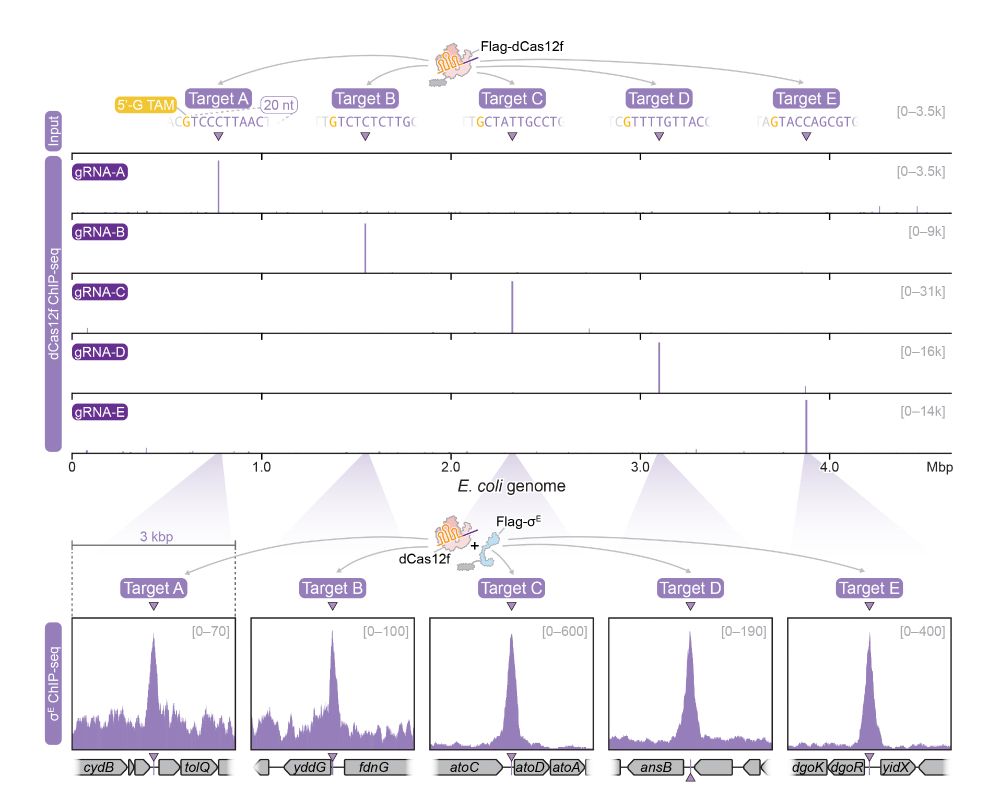
5/10 Indeed, when Cas12f is programmed to target various sites in the E. coli genome, we also observe recruitment of the Sigma factor to the same sites. dCas12f and Sigma appear to form a co-complex! Spoiler: check out our companion structure article with the Chang Lab!
June 11, 2025 at 4:03 PM
5/10 Indeed, when Cas12f is programmed to target various sites in the E. coli genome, we also observe recruitment of the Sigma factor to the same sites. dCas12f and Sigma appear to form a co-complex! Spoiler: check out our companion structure article with the Chang Lab!
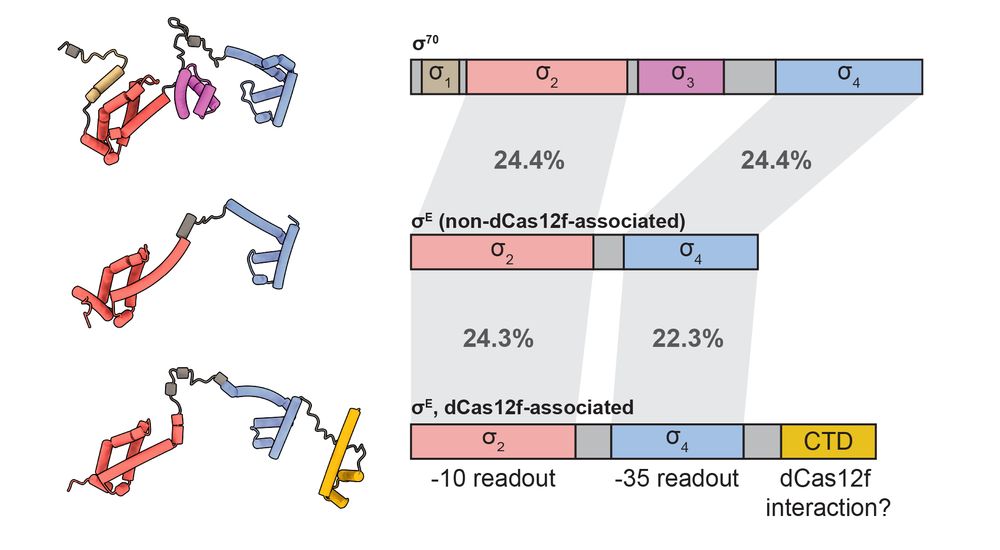
4/10 Sigma factors are essential for driving bacterial transcription by RNA polymerase (RNAP) from specific promoter sequence motifs. dCas12f-associated Sigma factors have an atypical, extended C-terminal domain, potentially facilitating interaction between Sigma and Cas12f.
June 11, 2025 at 4:03 PM
4/10 Sigma factors are essential for driving bacterial transcription by RNA polymerase (RNAP) from specific promoter sequence motifs. dCas12f-associated Sigma factors have an atypical, extended C-terminal domain, potentially facilitating interaction between Sigma and Cas12f.
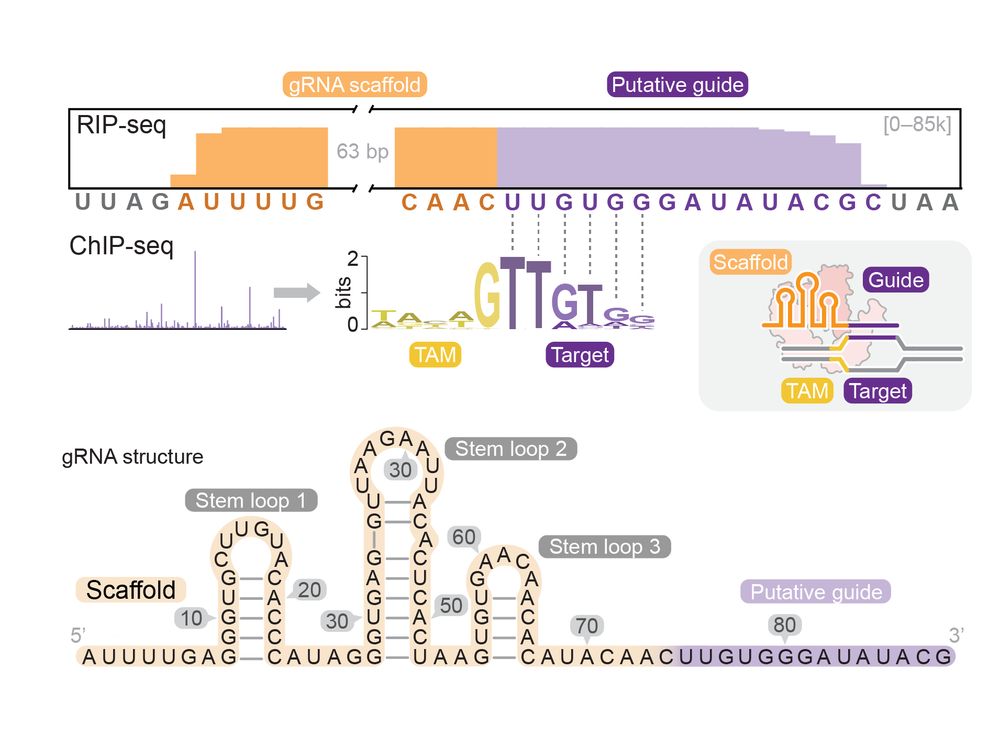
3/10 The dead-Cas12f is highly compact (~370 aa) and binds a natural single-guide RNA (~90 nt). Strikingly, dCas12f has minimal PAM/TAM requirements. It can target custom sites flanked by a simple 5’-G nucleotide.
June 11, 2025 at 4:03 PM
3/10 The dead-Cas12f is highly compact (~370 aa) and binds a natural single-guide RNA (~90 nt). Strikingly, dCas12f has minimal PAM/TAM requirements. It can target custom sites flanked by a simple 5’-G nucleotide.
2/10 Cas proteins typically defend against bacterial viruses by cleaving their genomes. The Cas12f homologs we identified in bacteria (see also Altae-Tran et al. 2023) are nuclease-dead (dCas12f) and have a conserved association with SigmaE factors (RpoE), suggesting a new function.
June 11, 2025 at 4:03 PM
2/10 Cas proteins typically defend against bacterial viruses by cleaving their genomes. The Cas12f homologs we identified in bacteria (see also Altae-Tran et al. 2023) are nuclease-dead (dCas12f) and have a conserved association with SigmaE factors (RpoE), suggesting a new function.
1/10 New pre-print(s) from the Sternberg Lab in collaboration with Leifu Chang's Lab! We uncover the unprecedented molecular mechanism of CRISPR-Cas12f-like proteins, which drive RNA-guided transcription independently of canonical promoter motifs.
Full story here:
www.biorxiv.org/content/10.1...
Full story here:
www.biorxiv.org/content/10.1...
June 11, 2025 at 4:03 PM
1/10 New pre-print(s) from the Sternberg Lab in collaboration with Leifu Chang's Lab! We uncover the unprecedented molecular mechanism of CRISPR-Cas12f-like proteins, which drive RNA-guided transcription independently of canonical promoter motifs.
Full story here:
www.biorxiv.org/content/10.1...
Full story here:
www.biorxiv.org/content/10.1...
"..one of the biggest challenges right now for evoCAST, and other large DNA editing tools under development, is delivery.
'How do we actually get these tools and their payloads into the cells or tissues of interest?'”
www.cuimc.columbia.edu/news/sternbe...
'How do we actually get these tools and their payloads into the cells or tissues of interest?'”
www.cuimc.columbia.edu/news/sternbe...

May 19, 2025 at 1:57 PM
"..one of the biggest challenges right now for evoCAST, and other large DNA editing tools under development, is delivery.
'How do we actually get these tools and their payloads into the cells or tissues of interest?'”
www.cuimc.columbia.edu/news/sternbe...
'How do we actually get these tools and their payloads into the cells or tissues of interest?'”
www.cuimc.columbia.edu/news/sternbe...
9/10 We establish evoCAST as a platform for efficient, programmable gene integration in human cells w/ potential for applications in therapeutics & research. We’re excited by the potential of CAST-PACE as a method for enhancing additional CAST systems that we continue to mine.
May 15, 2025 at 10:49 PM
9/10 We establish evoCAST as a platform for efficient, programmable gene integration in human cells w/ potential for applications in therapeutics & research. We’re excited by the potential of CAST-PACE as a method for enhancing additional CAST systems that we continue to mine.
8/10 evoCAST often achieved 10 to 30% integration efficiencies across 14 sites in human cells, an average 420-fold improvement over wild-type CAST 🤯🤯🤯

May 15, 2025 at 10:49 PM
8/10 evoCAST often achieved 10 to 30% integration efficiencies across 14 sites in human cells, an average 420-fold improvement over wild-type CAST 🤯🤯🤯
5/10 After performing hundreds of evolutionary generations in PACE, we generated an evolved variant of the CAST transposase protein TnsB with ten (!!) activity-enhancing mutations enabling >200-fold improved integration activity in human cells.

May 15, 2025 at 10:49 PM
5/10 After performing hundreds of evolutionary generations in PACE, we generated an evolved variant of the CAST transposase protein TnsB with ten (!!) activity-enhancing mutations enabling >200-fold improved integration activity in human cells.

