
Website: https://steglelab.org/
During her molecular biology studies, Gesa developed an interest in applying mathematics and ML to understand complex biological systems. Her PhD research will focus on inferring GRNs by integrating insights from population genetics and genetic perturbation data.

During her molecular biology studies, Gesa developed an interest in applying mathematics and ML to understand complex biological systems. Her PhD research will focus on inferring GRNs by integrating insights from population genetics and genetic perturbation data.
Steffi joins us as a PhD student after completing her Master’s at Heidelberg University. She will explore gene regulation and cell differentiation using organoid models and is excited to analyze these complex datasets and develop ML methods.

Steffi joins us as a PhD student after completing her Master’s at Heidelberg University. She will explore gene regulation and cell differentiation using organoid models and is excited to analyze these complex datasets and develop ML methods.



You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n

You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
Hakime shared reflections on the recently concluded Health Privacy Challenge and also organized a follow-up workshop. 🛡️

Hakime shared reflections on the recently concluded Health Privacy Challenge and also organized a follow-up workshop. 🛡️
Huge thanks to the organizers for such a fantastic event! 🙌

Huge thanks to the organizers for such a fantastic event! 🙌







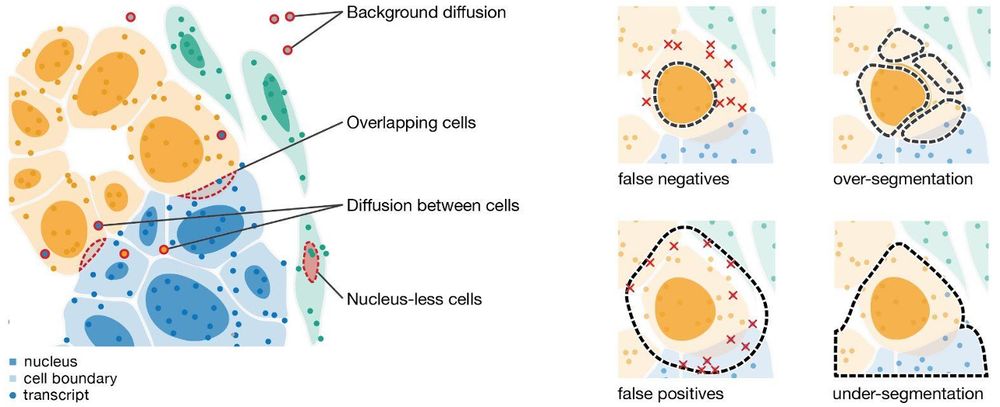
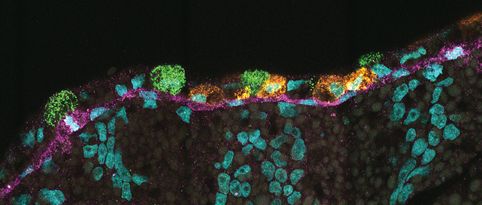
@elihei.bsky.social and our team at @embl.org , @dkfz.bsky.social, and @mskcancercenter.bsky.social built #segger - a fast, accurate cell segmentation tool for spatial transcriptomics that assigns transcripts to their cell origins!
doi.org/10.1101/2025...

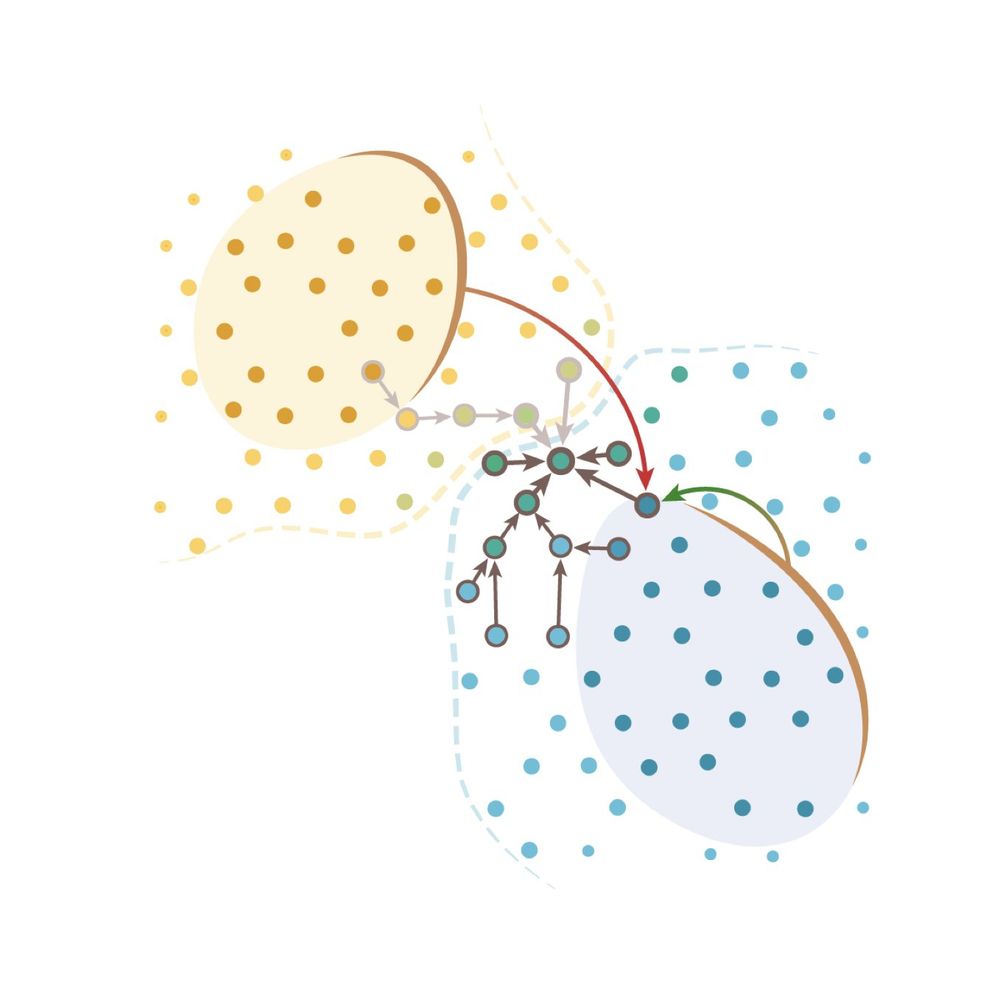
@elihei.bsky.social and our team at @embl.org , @dkfz.bsky.social, and @mskcancercenter.bsky.social built #segger - a fast, accurate cell segmentation tool for spatial transcriptomics that assigns transcripts to their cell origins!
doi.org/10.1101/2025...



1️⃣ Multi-omic profiling of medulloblastoma characterizes subclonal variation of chromothripsis (doi.org/10.1038/s414...)
2️⃣ Spatio-temporal profiling explores mechanisms of treatment resistance (doi.org/10.1038/s414...)
Thanks to all involved! 🙌

1️⃣ Multi-omic profiling of medulloblastoma characterizes subclonal variation of chromothripsis (doi.org/10.1038/s414...)
2️⃣ Spatio-temporal profiling explores mechanisms of treatment resistance (doi.org/10.1038/s414...)
Thanks to all involved! 🙌

