Babraham Institute and Enhanc3D Genomics









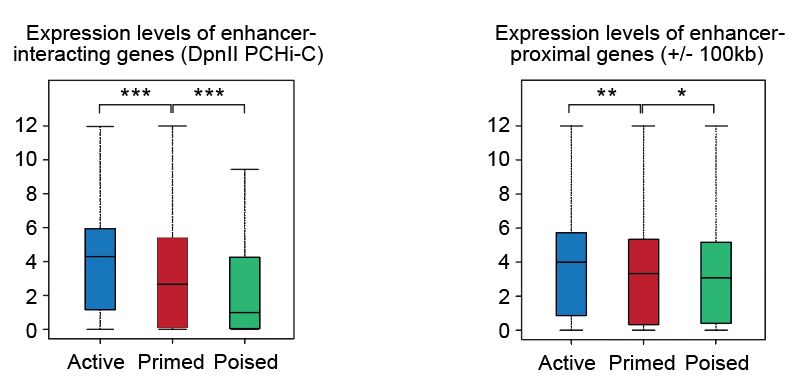
- Active (ATAC-seq+; H3K4me1+; H3K27ac+)
- Primed/Inactive (ATAC-seq+; H3K4me1+; H3K27ac-)
- Poised (ATAC-seq+; H3K27me3+)
- Chromatin repressed (ATAC-seq-)

- Active (ATAC-seq+; H3K4me1+; H3K27ac+)
- Primed/Inactive (ATAC-seq+; H3K4me1+; H3K27ac-)
- Poised (ATAC-seq+; H3K27me3+)
- Chromatin repressed (ATAC-seq-)
Join us at:
us02web.zoom.us/webinar/regi...

Join us at:
us02web.zoom.us/webinar/regi...
📅 March 13th 2025
⏰ 4 pm GMT
Join us at: us02web.zoom.us/webinar/regi...

📅 March 13th 2025
⏰ 4 pm GMT
Join us at: us02web.zoom.us/webinar/regi...

