
So are immortalized cells “damaged”? Not quite.
They’re not perfect stand-ins, but they’re surprisingly consistent with primary cells.
We conclude that Jurkat cells are a solid model for studying ψ — just watch out for that 13%.
📖 tinyurl.com/TCellPsiRNA
So are immortalized cells “damaged”? Not quite.
They’re not perfect stand-ins, but they’re surprisingly consistent with primary cells.
We conclude that Jurkat cells are a solid model for studying ψ — just watch out for that 13%.
📖 tinyurl.com/TCellPsiRNA
The twist? It’s not due to differences in pseudouridine synthase expression — enzyme levels were similar.
So what's driving site selection? Likely trans-regulatory factors or RNA structure, not just enzyme abundance.
The twist? It’s not due to differences in pseudouridine synthase expression — enzyme levels were similar.
So what's driving site selection? Likely trans-regulatory factors or RNA structure, not just enzyme abundance.
☑️ Jurkat-specific ψ-sites hit oncogenic and immune activation genes
☑️ Primary T cell–specific ψ-sites appear on trafficking and calcium signaling genes
🧬 Immortalized cells also showed more clustered ψ patterns, hinting at altered control compared to primary T cells.

☑️ Jurkat-specific ψ-sites hit oncogenic and immune activation genes
☑️ Primary T cell–specific ψ-sites appear on trafficking and calcium signaling genes
🧬 Immortalized cells also showed more clustered ψ patterns, hinting at altered control compared to primary T cells.
Most differences came from gene expression, not ψ-site selection.
This process seems to be tightly conserved.

Most differences came from gene expression, not ψ-site selection.
This process seems to be tightly conserved.
Huge thanks to the team for all their hard work! @wanunupore.bsky.social @genometdcc.bsky.social
Huge thanks to the team for all their hard work! @wanunupore.bsky.social @genometdcc.bsky.social
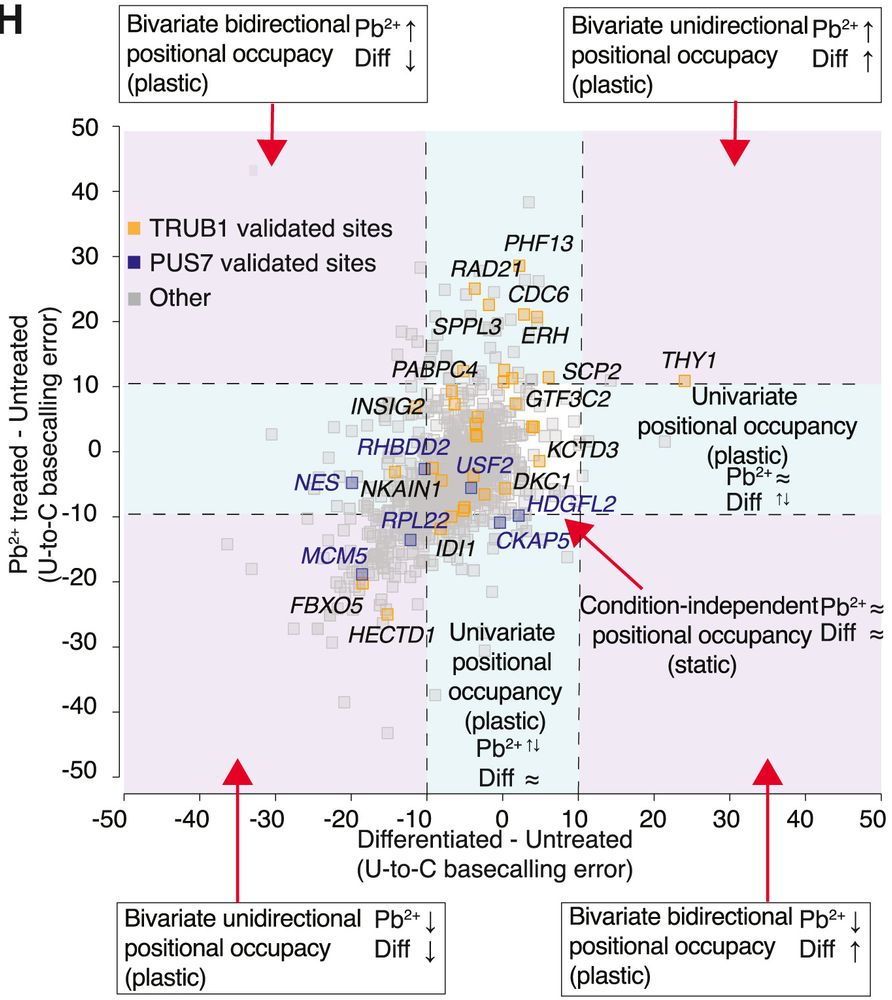
Method: SH SY5Y cells were untreated, differentiated with retinoic acid, or exposed to Pb²⁺. Nanopore DRS determined site-specific, relative Ψ “occupancy”; orthogonal knockdowns (TRUB1, PUS7) and biochemical assays validated sites.
Method: SH SY5Y cells were untreated, differentiated with retinoic acid, or exposed to Pb²⁺. Nanopore DRS determined site-specific, relative Ψ “occupancy”; orthogonal knockdowns (TRUB1, PUS7) and biochemical assays validated sites.

