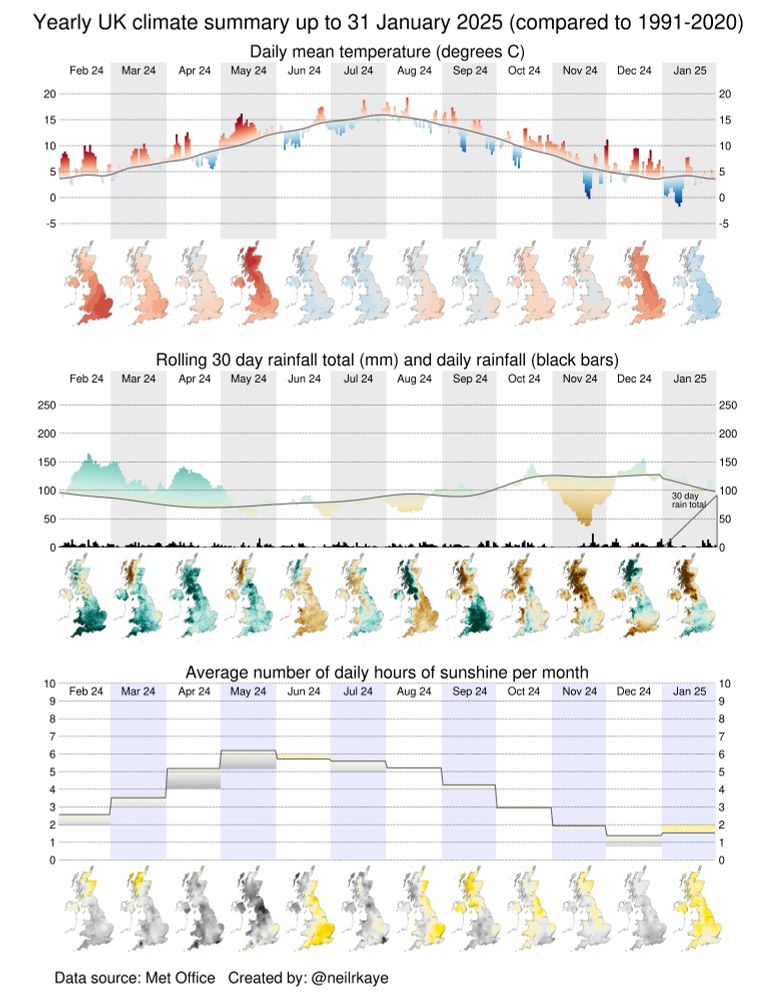
Save the plot once 👌
e.g.:
ggplot(...) +
ggview::canvas(width = 220, height = 220*2/3, units = "mm", dpi = 300)
per @nrennie.bsky.social #datavis #rstats
Save the plot once 👌
e.g.:
ggplot(...) +
ggview::canvas(width = 220, height = 220*2/3, units = "mm", dpi = 300)
per @nrennie.bsky.social #datavis #rstats
You can run your package unit tests with {testthat} in parallel with two simple steps (see testthat.r-lib.org/articles/par...):
tldr:
1. Add `Config/testthat/parallel: true` to DESCRIPTION.
2. Add `TESTTHAT_CPUS=8` to your .Renviron and restart R.
#rstats
You can run your package unit tests with {testthat} in parallel with two simple steps (see testthat.r-lib.org/articles/par...):
tldr:
1. Add `Config/testthat/parallel: true` to DESCRIPTION.
2. Add `TESTTHAT_CPUS=8` to your .Renviron and restart R.
#rstats
#rstats
#rstats
medium.com/p/3aa0b039410
(Post 4 out of 4 while I'm on holiday, sharing some highlights from previous years)

medium.com/p/3aa0b039410
(Post 4 out of 4 while I'm on holiday, sharing some highlights from previous years)
histogram_guide <- compose_sandwich(
middle = gizmo_histogram(just = 0),
text = "axis_base",
position = "bottom"
)
nc <- sf::st_read(system.file("shape/nc.shp", package = "sf"), quiet = TRUE)
ggplot(nc) +
geom_sf(aes(fill = AREA)) +
guides(fill = histogram_guide)

histogram_guide <- compose_sandwich(
middle = gizmo_histogram(just = 0),
text = "axis_base",
position = "bottom"
)
nc <- sf::st_read(system.file("shape/nc.shp", package = "sf"), quiet = TRUE)
ggplot(nc) +
geom_sf(aes(fill = AREA)) +
guides(fill = histogram_guide)