Great collaboration between BOKU University, IMBA, MedUni Vienna, and Michigan University! Thank you, J. Helm and the amazing Stadlmann Lab for taking me on this ride!! Together, we’re uncovering new insights into glycans. 🟦 🟡 🔺 🟢 🔶 🟨
Great collaboration between BOKU University, IMBA, MedUni Vienna, and Michigan University! Thank you, J. Helm and the amazing Stadlmann Lab for taking me on this ride!! Together, we’re uncovering new insights into glycans. 🟦 🟡 🔺 🟢 🔶 🟨
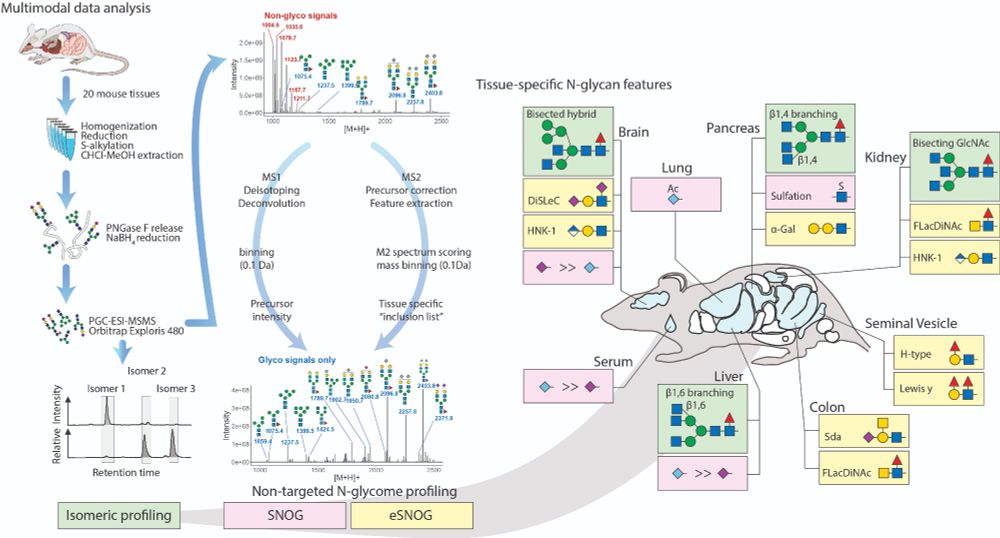
In our paper, we showcase a scalable, LC-MS/MS data-driven method combining automated ID, non-targeted profiling, and isomer-sensitive glycan analysis. Dive into the details: doi.org/10.1038/s414...
In our paper, we showcase a scalable, LC-MS/MS data-driven method combining automated ID, non-targeted profiling, and isomer-sensitive glycan analysis. Dive into the details: doi.org/10.1038/s414...
For those ready to dive deeper: We haven’t even started with the chromatography data! PGC allows us to differentiate N-glycans with the same mass but distinct structures, adding more layers of tissue-specific complexity.
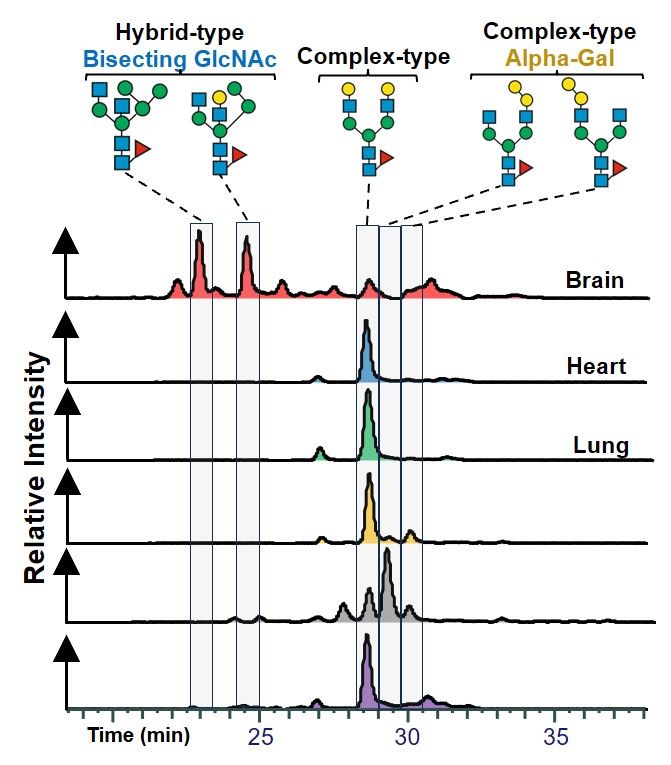
For those ready to dive deeper: We haven’t even started with the chromatography data! PGC allows us to differentiate N-glycans with the same mass but distinct structures, adding more layers of tissue-specific complexity.
But wait—there was more! After filtering non-glycan spectra and expected glycans, we found spectra representing unexpected glycan structures and mapped them across all tissues! 🔍
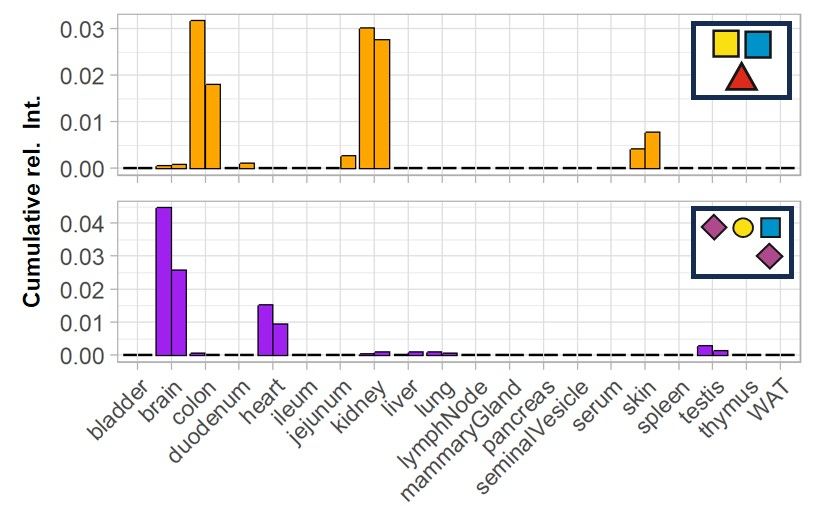
But wait—there was more! After filtering non-glycan spectra and expected glycans, we found spectra representing unexpected glycan structures and mapped them across all tissues! 🔍
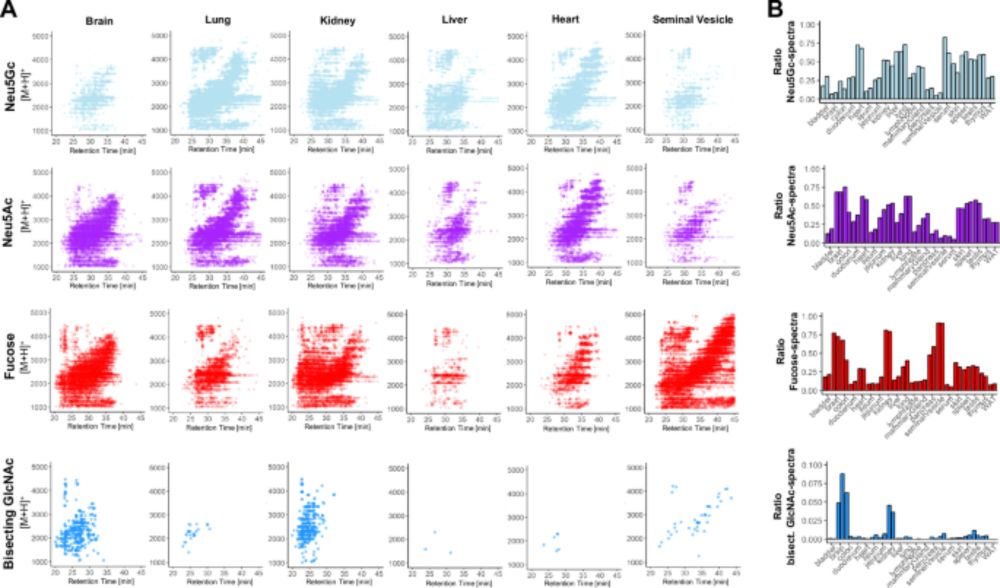
Next, we extended SNOG with eSNOG, extracting spectra with special fragment ions to compare glycan structural features like α-Gal, antennary Fuc, Neu5Ac, Neu5Gc, and oligo-Man across tissues.
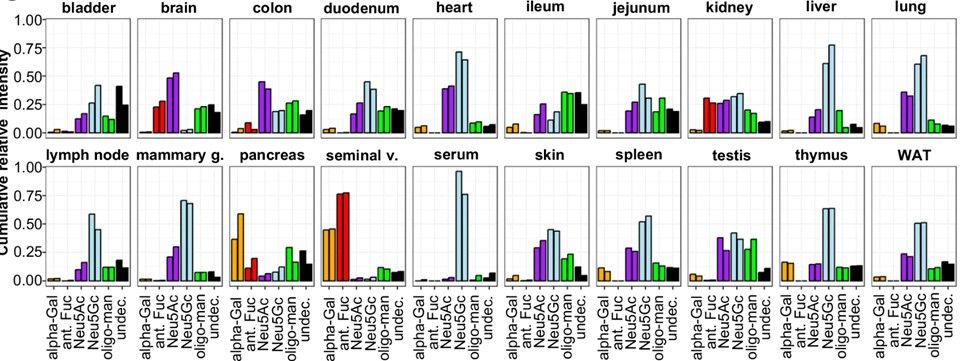
Next, we extended SNOG with eSNOG, extracting spectra with special fragment ions to compare glycan structural features like α-Gal, antennary Fuc, Neu5Ac, Neu5Gc, and oligo-Man across tissues.
With SNOG-filtered data, we saw clear, tissue-specific glycan clusters emerge, with high technical reproducibility across replicates. This filtering step was a game-changer!
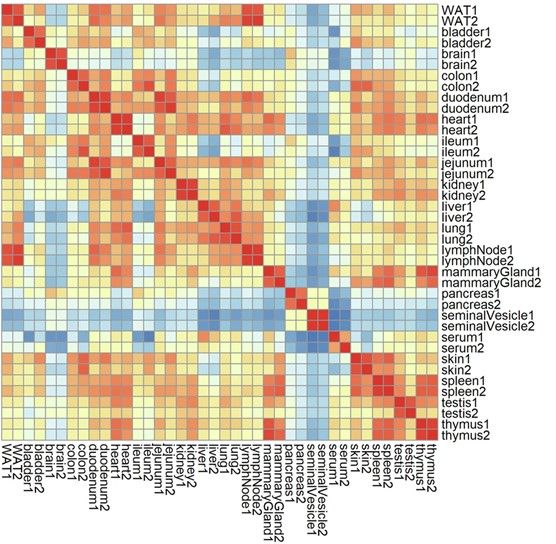
With SNOG-filtered data, we saw clear, tissue-specific glycan clusters emerge, with high technical reproducibility across replicates. This filtering step was a game-changer!
First up: SNOG-filtering. This automated step removes contaminants (up to 75% in some tissues!) so we can focus on genuine N-glycan signals.
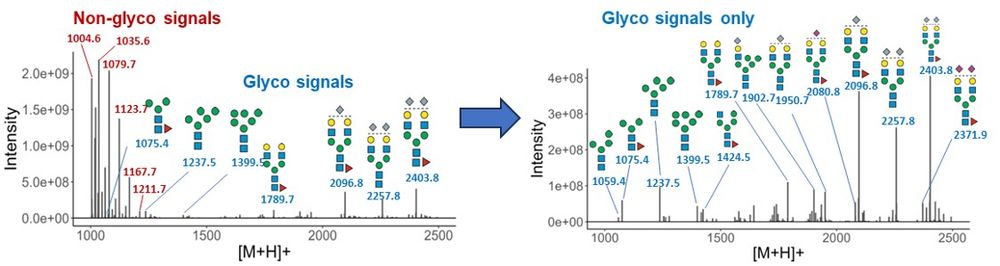
First up: SNOG-filtering. This automated step removes contaminants (up to 75% in some tissues!) so we can focus on genuine N-glycan signals.
Our approach had to be scalable, non-targeted (no pre-built mass lists, please!), and data-driven. Automated spectral ID would be the icing on the cake! 🧁 We combined PGC analysis with multiple profiling strategies for semi/quantitative N-glycomics.

Our approach had to be scalable, non-targeted (no pre-built mass lists, please!), and data-driven. Automated spectral ID would be the icing on the cake! 🧁 We combined PGC analysis with multiple profiling strategies for semi/quantitative N-glycomics.
Faced with an overwhelming amount of LC-MS/MS data, we knew manual annotation was out of the question. Instead, we developed an “unconventional” approach to analyze our N-glycome data in a faster, smarter way. Here’s what we did:
Faced with an overwhelming amount of LC-MS/MS data, we knew manual annotation was out of the question. Instead, we developed an “unconventional” approach to analyze our N-glycome data in a faster, smarter way. Here’s what we did:
So, we analyzed N-glycans from 20 mouse tissues, generating a comprehensive, structure-resolved N-glycome Atlas. Using PGC columns for isomer separation and an Orbitrap for high-res MS/MS, we assembled a mountain of data. Now what?

So, we analyzed N-glycans from 20 mouse tissues, generating a comprehensive, structure-resolved N-glycome Atlas. Using PGC columns for isomer separation and an Orbitrap for high-res MS/MS, we assembled a mountain of data. Now what?
Have you ever wondered how many unique N-glycan structures a single organism might create? Or if the sugars on your kidney cells differ from those in your brain?
We did too, and we set out to investigate!
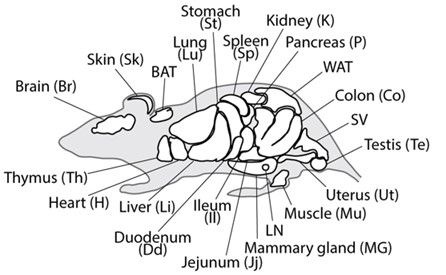
Have you ever wondered how many unique N-glycan structures a single organism might create? Or if the sugars on your kidney cells differ from those in your brain?
We did too, and we set out to investigate!

