Web: https://slavovlab.net
Videos: http://youtube.slavovlab.net
Multiple TFs transmit mechanical forces into transcriptional regulation.
Nuclear shape, size, and chromatin abnormalities are common features of many diseases.

Multiple TFs transmit mechanical forces into transcriptional regulation.
Nuclear shape, size, and chromatin abnormalities are common features of many diseases.

If important, make it count.
Make it easy to reproduce.
Share the data.
Share the code.
Share the metadata.
Use community guidelines to facilitate reanalysis & reuse of your results.
nature.com/articles/s41...

If important, make it count.
Make it easy to reproduce.
Share the data.
Share the code.
Share the metadata.
Use community guidelines to facilitate reanalysis & reuse of your results.
nature.com/articles/s41...
This study reported remarkably low correlations in the context of heat shock response and survival.

This study reported remarkably low correlations in the context of heat shock response and survival.
This explains the degree to which protein clearance (degradation) sets protein abundance.
biorxiv.org/content/10.1...

This explains the degree to which protein clearance (degradation) sets protein abundance.
biorxiv.org/content/10.1...
The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules

The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules
- Direct measurements
- Quantitative accuracy supporting confident modeling
- Scalable multiplexing & generalizable approach approach

- Direct measurements
- Quantitative accuracy supporting confident modeling
- Scalable multiplexing & generalizable approach approach
The results revealed 𝐜𝐨𝐦𝐩𝐥𝐞𝐱 regulation & 𝐬𝐢𝐦𝐩𝐥𝐞 organizing principles:
www.biorxiv.org/content/10.1...
🧵

The results revealed 𝐜𝐨𝐦𝐩𝐥𝐞𝐱 regulation & 𝐬𝐢𝐦𝐩𝐥𝐞 organizing principles:
www.biorxiv.org/content/10.1...
🧵
Below are some of the key regulators of mitosis.

Below are some of the key regulators of mitosis.
It's elegantly dynamic !
Proteins constantly self-assemble and disassemble to build skeletal structures in our cells, shaping and orchestrating life.

It's elegantly dynamic !
Proteins constantly self-assemble and disassemble to build skeletal structures in our cells, shaping and orchestrating life.
A decade of pushing the boundaries in proteomics, systems biology, and data-driven discovery.
🔬 Thank you to our collaborators, alumni & supporters!

A decade of pushing the boundaries in proteomics, systems biology, and data-driven discovery.
🔬 Thank you to our collaborators, alumni & supporters!
In close range (below 1kb), the knockout affects the abundance of mRNA transcribed from nearby genes.
Yeast Data 🔽

In close range (below 1kb), the knockout affects the abundance of mRNA transcribed from nearby genes.
Yeast Data 🔽
This is a crucial factor to consider in knockout experiments.
Phenotypic similarity is exponentially related to chromosomal proximity in both yeast & human genomes.
Human Data 🔽

This is a crucial factor to consider in knockout experiments.
Phenotypic similarity is exponentially related to chromosomal proximity in both yeast & human genomes.
Human Data 🔽

During splicing, stepwise assembly is one principle by which spliceosomes on the one hand ensure the faithful recognition of splice sites and on the other hand become susceptible to regulatory inputs to implement alternative splicing.

During splicing, stepwise assembly is one principle by which spliceosomes on the one hand ensure the faithful recognition of splice sites and on the other hand become susceptible to regulatory inputs to implement alternative splicing.

A creative use of the impressive imaging capabilities developed for DNA sequencing by synthesis.
◼️ It allowed quantifying binding and enzyme catalysis
🧵

A creative use of the impressive imaging capabilities developed for DNA sequencing by synthesis.
◼️ It allowed quantifying binding and enzyme catalysis
🧵
The mechanisms that generate and maintain this extraordinary diversity of sizes remain incompletely understood.

The mechanisms that generate and maintain this extraordinary diversity of sizes remain incompletely understood.
It can analyze the proteomes of ~ 1,000 single cells / day, and we need to ensure high quality cell isolation and sample preparation:
⬛️ These approaches can help:
nature.com/articles/s41...

It can analyze the proteomes of ~ 1,000 single cells / day, and we need to ensure high quality cell isolation and sample preparation:
⬛️ These approaches can help:
nature.com/articles/s41...
Tree representations for distortion-free visualization and exploratory analysis of single-cell omics data.
The trees are constructed to accurately represent true distances between the objects in the high-dimensional space.
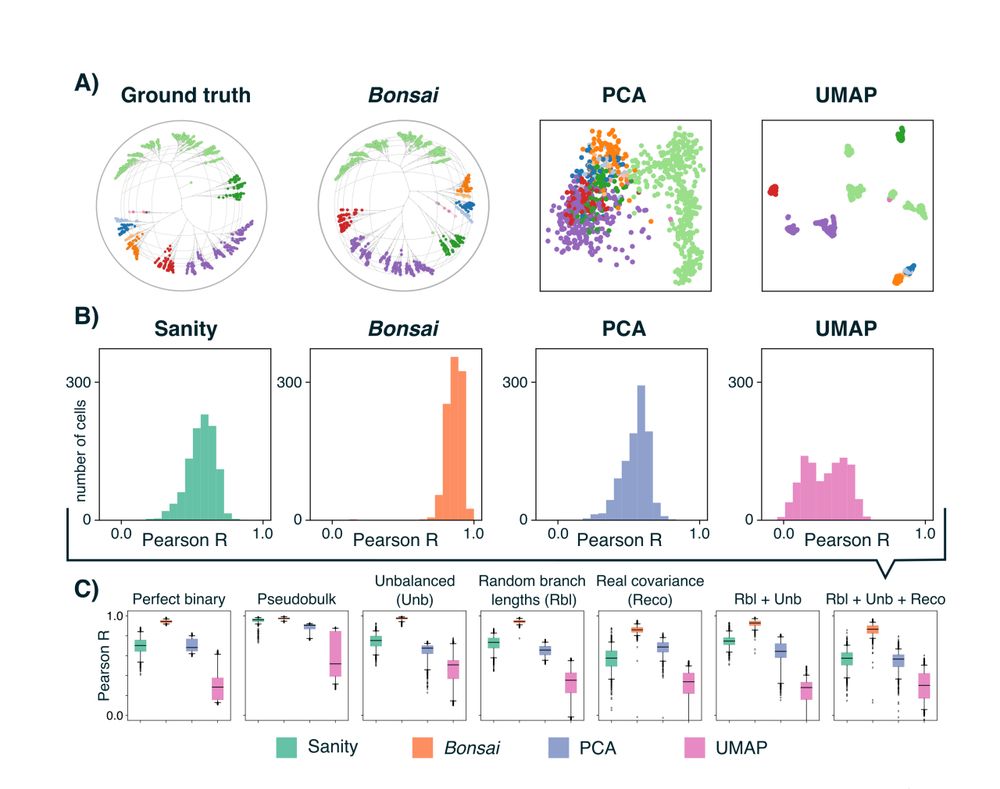
Tree representations for distortion-free visualization and exploratory analysis of single-cell omics data.
The trees are constructed to accurately represent true distances between the objects in the high-dimensional space.


Why is such a large complex needed for a simple cleavage ?
We show how specific protein subunits distinguish RNase MRP from related enzymes and define its catalytic repertoar.
www.biorxiv.org/content/10.1...

Why is such a large complex needed for a simple cleavage ?
We show how specific protein subunits distinguish RNase MRP from related enzymes and define its catalytic repertoar.
www.biorxiv.org/content/10.1...

Protein degradation is a HUGE factor.
It accounts for up to 50 % of protein variation across proteins & tissue types.
www.biorxiv.org/content/10.1...
🧵
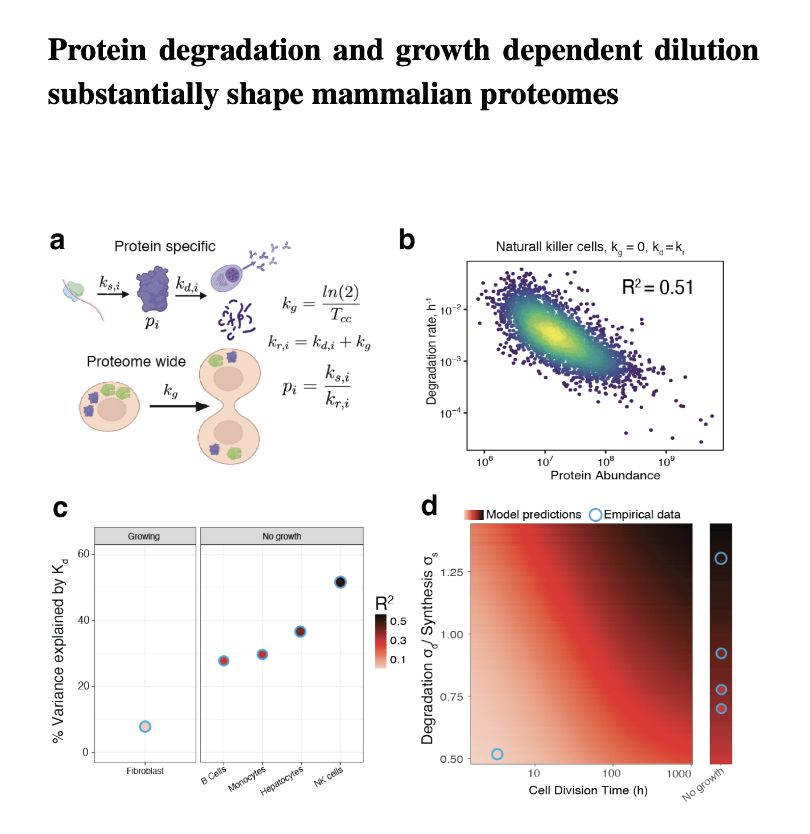
Protein degradation is a HUGE factor.
It accounts for up to 50 % of protein variation across proteins & tissue types.
www.biorxiv.org/content/10.1...
🧵

