
Focused on quantitative disease resistance
Passioned about system-sciences
Agronomist @CAU





We conducted a #phylotranscriptomic study on our most favourite Solanum species:

We conducted a #phylotranscriptomic study on our most favourite Solanum species:



Interestingly, the resistant genotype shows constitutively high WRKY6 expression. Almost as if it's primed.
7/n
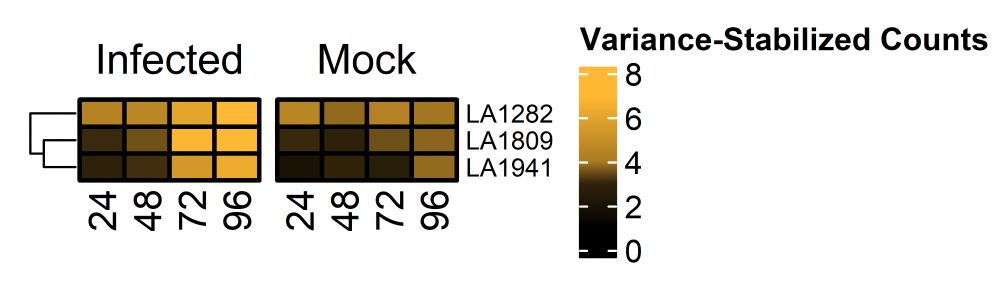
Interestingly, the resistant genotype shows constitutively high WRKY6 expression. Almost as if it's primed.
7/n
Clustering based on Bayesian and Dirichlet statistics revealed that key genes were expressed earlier in the resistant genotype and delayed in the susceptible ones!
6/n
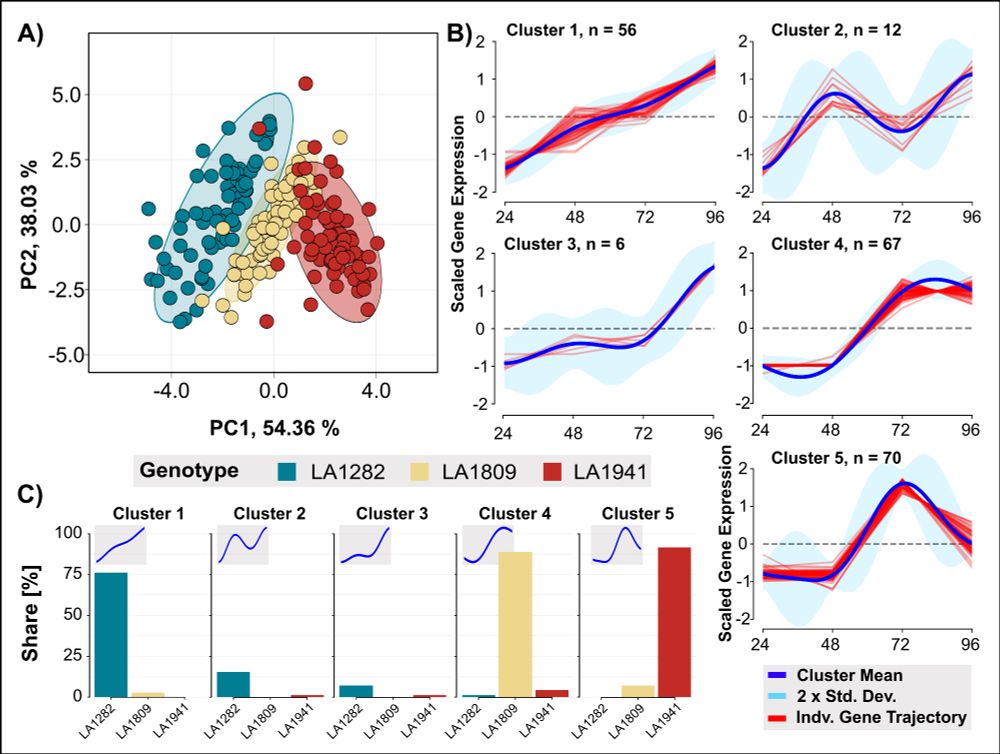
Clustering based on Bayesian and Dirichlet statistics revealed that key genes were expressed earlier in the resistant genotype and delayed in the susceptible ones!
6/n
These included classic pathogenesis-related genes like PR1 and PR4, as well as genes involved in oxidative stress responses.
5/n
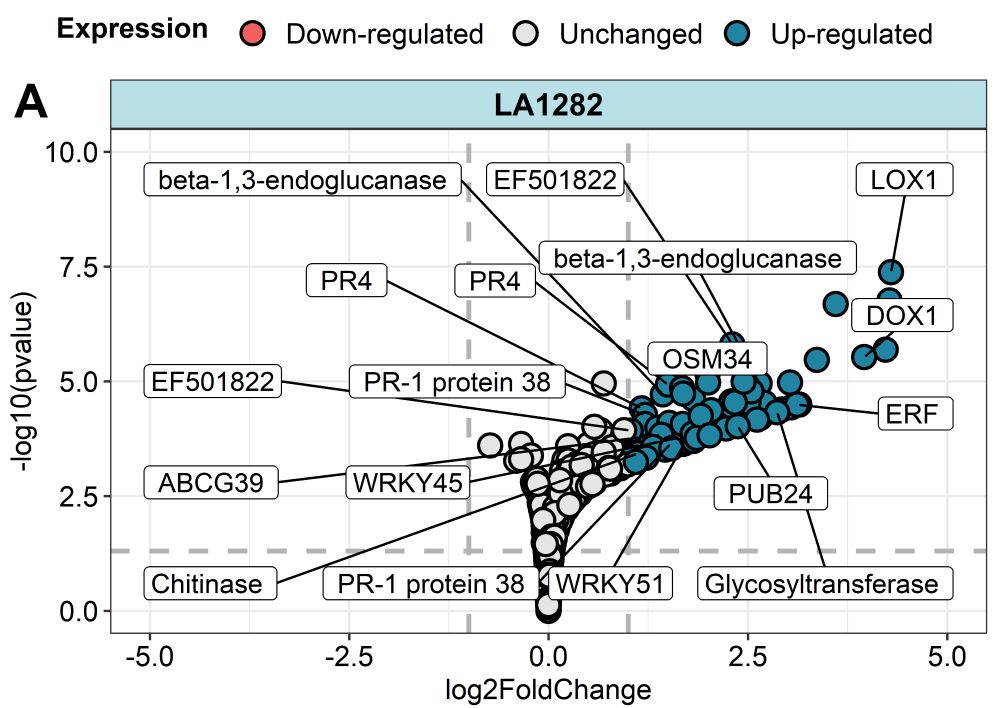
These included classic pathogenesis-related genes like PR1 and PR4, as well as genes involved in oxidative stress responses.
5/n
Interestingly, we observed relatively little early infection-induced gene expression: most strong differential expression occurred only after infection was well established.
4/n
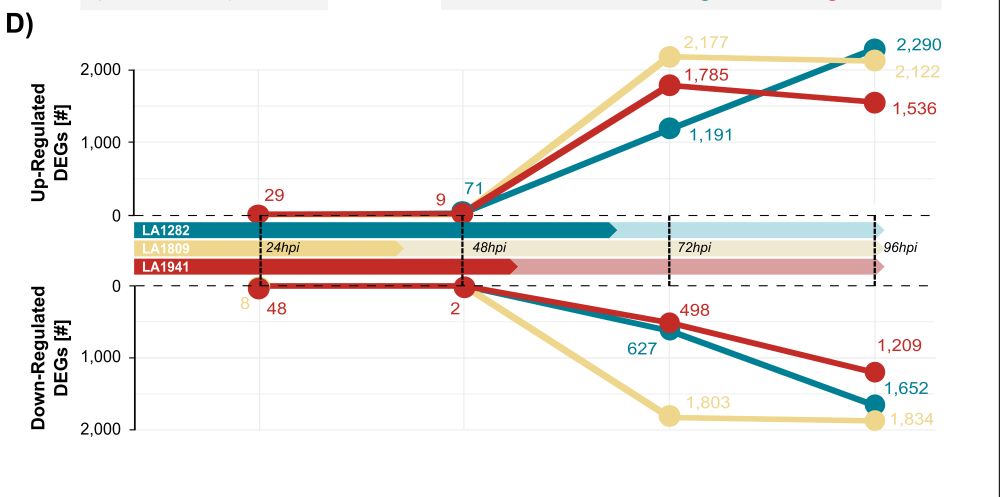
Interestingly, we observed relatively little early infection-induced gene expression: most strong differential expression occurred only after infection was well established.
4/n
Using the PhenoVation PlantExplorer Pro, we validated lag phase duration phenotypes by tracking Fv/Fm as a proxy for photosynthetic stress.
This allowed us to pinpoint early physiological changes between genotypes.
3/n
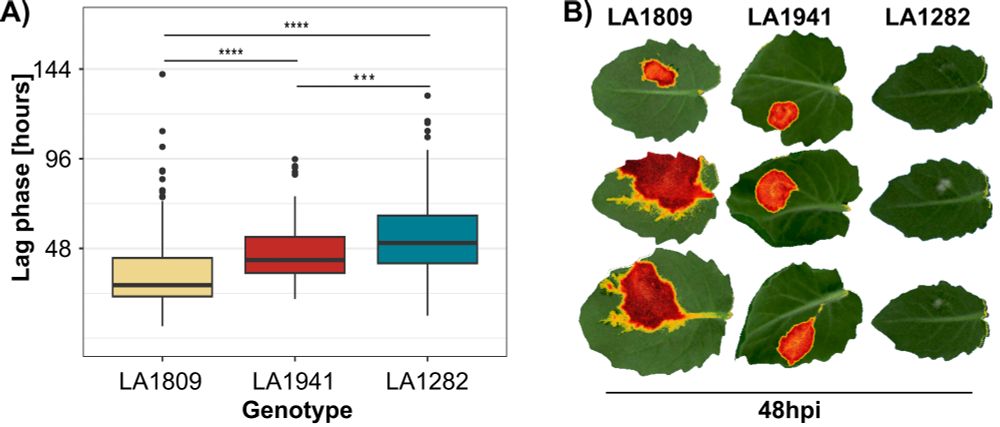
Using the PhenoVation PlantExplorer Pro, we validated lag phase duration phenotypes by tracking Fv/Fm as a proxy for photosynthetic stress.
This allowed us to pinpoint early physiological changes between genotypes.
3/n
I’m deeply grateful for the incredible support and mentoring from @rstam.bsky.social and for my amazing colleagues, family, and friends!

I’m deeply grateful for the incredible support and mentoring from @rstam.bsky.social and for my amazing colleagues, family, and friends!
all S. chilense accessions survived a super windy & rainy weekend: not a single loss!
tough wild tomatoes. they’re even thriving
#plantscience #StressTolerance #Solanaceae #SciComm

all S. chilense accessions survived a super windy & rainy weekend: not a single loss!
tough wild tomatoes. they’re even thriving
#plantscience #StressTolerance #Solanaceae #SciComm
We’ve just started a field trial with Solanum chilense at the @uni-kiel.de Botanical Garden.
Can’t wait to watch our wild tomatoes tackle the Northern German summer. 🍅
Stay tuned for our next field-trial updates!
#SChilense #FieldTrial #PlantScience #Kiel


We’ve just started a field trial with Solanum chilense at the @uni-kiel.de Botanical Garden.
Can’t wait to watch our wild tomatoes tackle the Northern German summer. 🍅
Stay tuned for our next field-trial updates!
#SChilense #FieldTrial #PlantScience #Kiel
Special thanks belongs to the whole AG @rstam.bsky.social for making this journey such a joy🍅🌿
Stay tuned for more exciting science on #tomato #resistance to be published! 🚀

Special thanks belongs to the whole AG @rstam.bsky.social for making this journey such a joy🍅🌿
Stay tuned for more exciting science on #tomato #resistance to be published! 🚀
Our second (and last) lab move to our new building begins!
Looking forward to state of the art lab spaces 🚀🍅🌿

Our second (and last) lab move to our new building begins!
Looking forward to state of the art lab spaces 🚀🍅🌿

