Computational and Systems Biology
Proteins and post-translational modifications
The buffering is much stronger than at the RNA level.

The buffering is much stronger than at the RNA level.
www.nature.com/articles/s41...
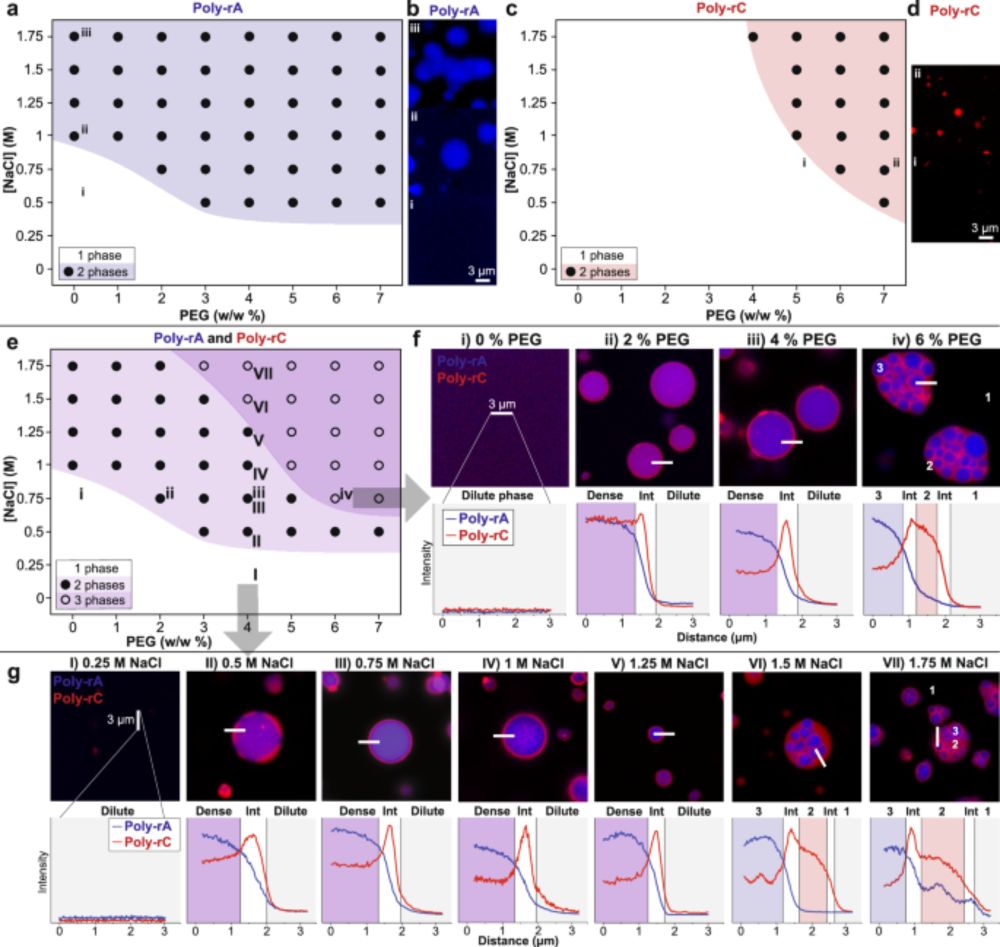
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
1/2
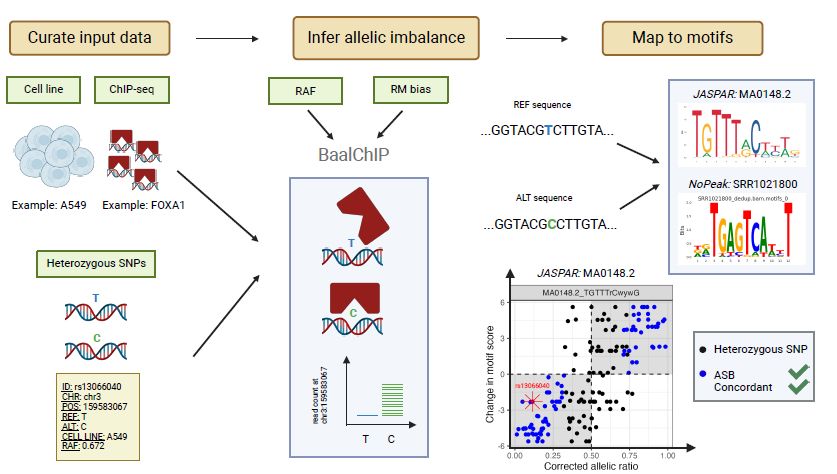
www.biorxiv.org/content/10.1...
1/2
www.biorxiv.org/content/10.1...
🧵👇 (1/n)

www.biorxiv.org/content/10.1...
🧵👇 (1/n)
www.biorxiv.org/content/10.1...
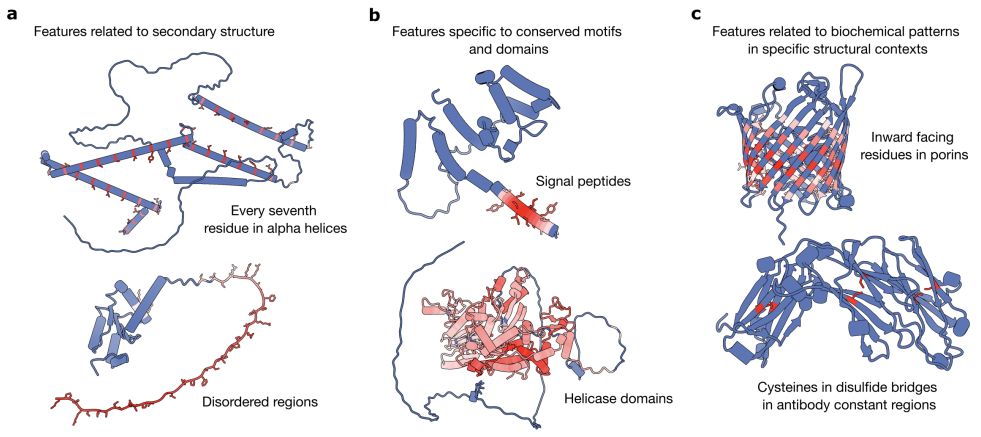
www.biorxiv.org/content/10.1...
Software: github.com/KosinskiLab/...
Preprint: biorxiv.org/content/10.1...


Software: github.com/KosinskiLab/...
Preprint: biorxiv.org/content/10.1...
They are stable & abundant O( 10³ ) copies / cell.
Generative mechanisms include codon-anticodon mismatches & RNA modifications.
Their abundance depends on codon frequency & protein stability.
www.biorxiv.org/content/10.1...

They are stable & abundant O( 10³ ) copies / cell.
Generative mechanisms include codon-anticodon mismatches & RNA modifications.
Their abundance depends on codon frequency & protein stability.
www.biorxiv.org/content/10.1...
Very proud of this work, thanks to a wonderful collaboration with Forest White lab at MIT, Cantley lab at DFCI, and leb by the amazingly talented @alissandra-hillis.bsky.social! Published as fully @openaccess.bsky.social in PNAS.
www.pnas.org/doi/10.1073/...

Very proud of this work, thanks to a wonderful collaboration with Forest White lab at MIT, Cantley lab at DFCI, and leb by the amazingly talented @alissandra-hillis.bsky.social! Published as fully @openaccess.bsky.social in PNAS.
www.pnas.org/doi/10.1073/...

