More details about the recruitment process here: chandar-lab.github.io/join/

More details about the recruitment process here: chandar-lab.github.io/join/

github.com/chandar-lab/...

github.com/chandar-lab/...






🧵 More in thread:
🧵 More in thread:
Results? Still far off—o3-mini scored only 12 out of 25 even with extensive prompting! 🚧 #LLMs 6/n
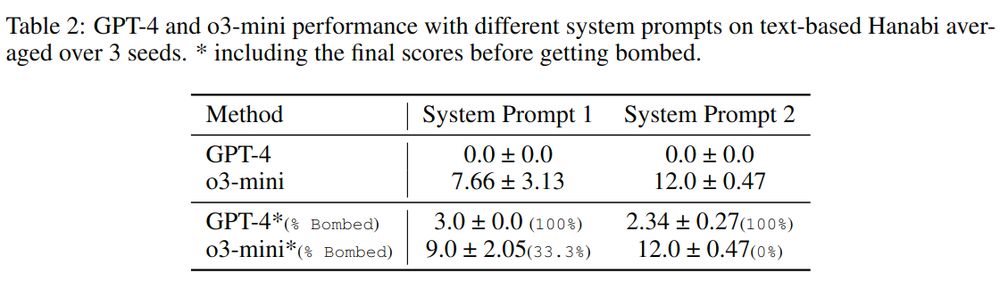
Results? Still far off—o3-mini scored only 12 out of 25 even with extensive prompting! 🚧 #LLMs 6/n
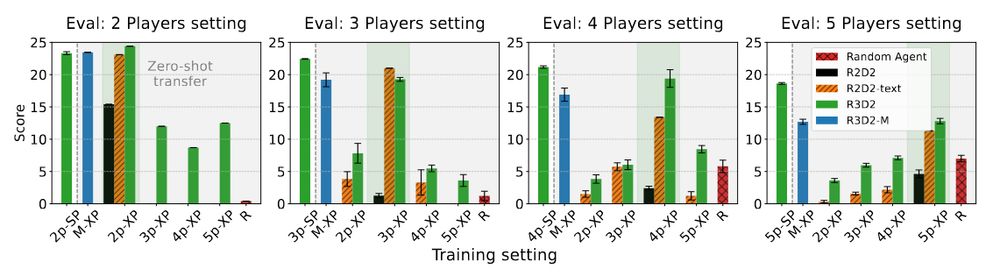
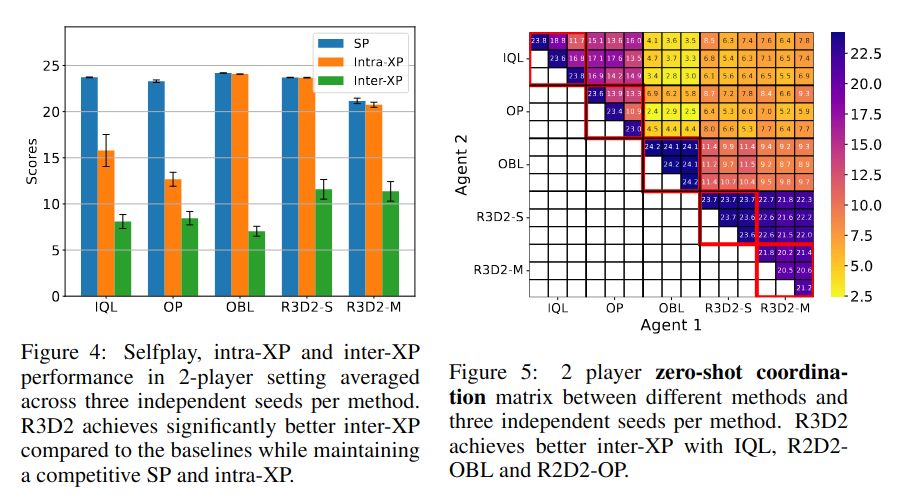

We explore this in our ICLR 2025 paper: A Generalist Hanabi Agent. We develop R3D2, the first agent to master all Hanabi settings and generalize to novel partners! 🚀 #ICLR2025 1/n
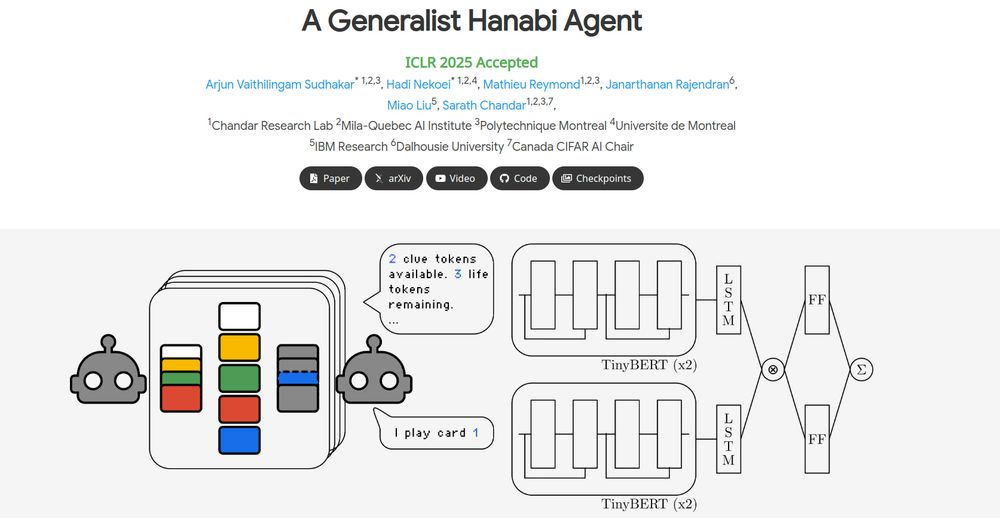
We explore this in our ICLR 2025 paper: A Generalist Hanabi Agent. We develop R3D2, the first agent to master all Hanabi settings and generalize to novel partners! 🚀 #ICLR2025 1/n

If you are interested in my spicy takes on ML for Biology, continue reading this thread! 1/n

If you are interested in my spicy takes on ML for Biology, continue reading this thread! 1/n