| Crop wild relatives and their pathogens | Likes quantitative and qualitative resistance mechanisms & popgen



Interestingly, Clade 1 has one unique Starship!
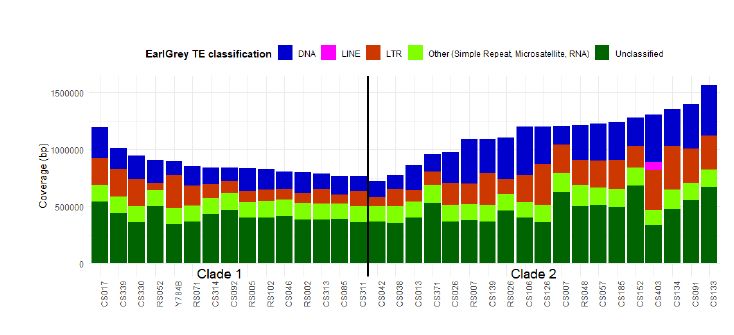
Interestingly, Clade 1 has one unique Starship!

We find some hints, but our study does not have the resolution to capture this in detail.
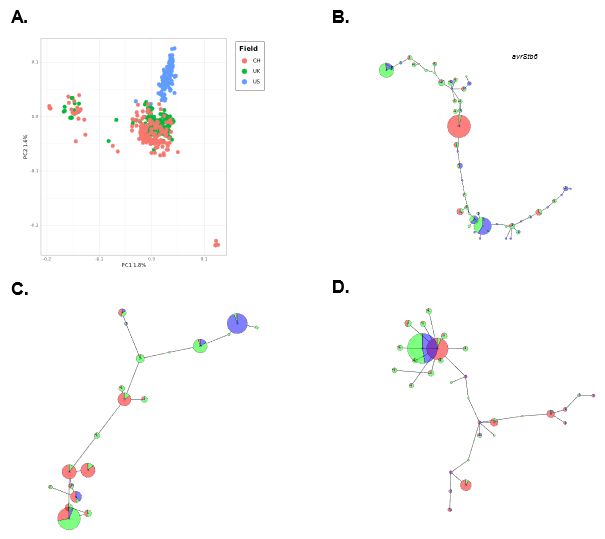
But we consistently find two genetic clades! Both infect Tomato throughout the range.

We find some hints, but our study does not have the resolution to capture this in detail.
But we consistently find two genetic clades! Both infect Tomato throughout the range.





The defences were excellent and they finished it off with the tradition of climbing the statue of meteor! de.m.wikipedia.org/wiki/Meteor_...


The defences were excellent and they finished it off with the tradition of climbing the statue of meteor! de.m.wikipedia.org/wiki/Meteor_...





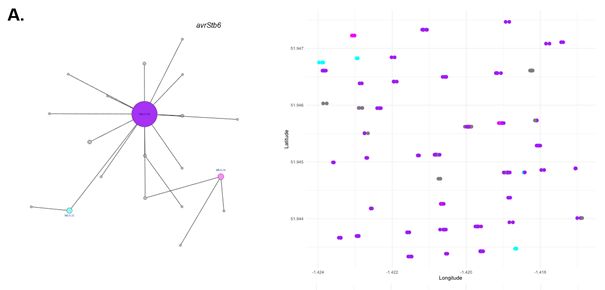
The US field population has lower number of SNPs. In UK and CH fields, SNP accumulation flattens slowly after collection 160+ isolates. Fields show some separation in a PCA.

The US field population has lower number of SNPs. In UK and CH fields, SNP accumulation flattens slowly after collection 160+ isolates. Fields show some separation in a PCA.
I'm looking forward to the defence (and some serious celebrations afterwards!) 🍾🥂

I'm looking forward to the defence (and some serious celebrations afterwards!) 🍾🥂
E.In four species, as in literature, this gene shows no role in defence. S. pennellii we see it central in the network and expression correlates with resistance

E.In four species, as in literature, this gene shows no role in defence. S. pennellii we see it central in the network and expression correlates with resistance




What better way to start the weekend than with a drink and an amazing story about Pseudomonas evolution and Kiwi fruit infection?


What better way to start the weekend than with a drink and an amazing story about Pseudomonas evolution and Kiwi fruit infection?