Rowan Green
@rowancallumg.bsky.social
Postdoc studying viral evolution with @asherleeks.bsky.social | previously @mermanchester.bsky.social | mutagenesis | bacteria | viruses | big R nerd 🧬🦠💻
🏳️⚧️ He/Him
🏳️⚧️ He/Him
Across domains of life 2017: doi.org/cb9s
Potential mechanism 2024: doi.org/m8zt
As discussed in the 2024 paper, ecological relevance likely depends on the amount of through-flow in the environment. Less flow = more opportunity for collective detoxification of the environment
Potential mechanism 2024: doi.org/m8zt
As discussed in the 2024 paper, ecological relevance likely depends on the amount of through-flow in the environment. Less flow = more opportunity for collective detoxification of the environment

Spontaneous mutation rate is a plastic trait associated with population density across domains of life
Author summary Spontaneous mutations fuel evolution, but the rate at which they occur can vary for a particular organism depending on its environment—a phenomenon known as mutation-rate plasticity. Fo...
doi.org
August 18, 2025 at 5:43 PM
Across domains of life 2017: doi.org/cb9s
Potential mechanism 2024: doi.org/m8zt
As discussed in the 2024 paper, ecological relevance likely depends on the amount of through-flow in the environment. Less flow = more opportunity for collective detoxification of the environment
Potential mechanism 2024: doi.org/m8zt
As discussed in the 2024 paper, ecological relevance likely depends on the amount of through-flow in the environment. Less flow = more opportunity for collective detoxification of the environment
Finally, a shoutout to the blueycolors colour scheme for making all of my ggplots far more fun! 🟦 🟧 13/13

August 18, 2025 at 3:56 PM
Finally, a shoutout to the blueycolors colour scheme for making all of my ggplots far more fun! 🟦 🟧 13/13
Deleting the nudJ gene not only makes rifampicin resistance less likely by reducing the mutation rate but also reduces the chance of a resistant mutant rising to fixation as ΔnudJ favours lower fitness mutations: synergystic effects of mutation rates and spectra impeding resistance evolution 12/13
August 18, 2025 at 3:54 PM
Deleting the nudJ gene not only makes rifampicin resistance less likely by reducing the mutation rate but also reduces the chance of a resistant mutant rising to fixation as ΔnudJ favours lower fitness mutations: synergystic effects of mutation rates and spectra impeding resistance evolution 12/13
Resistance mutations accessed by ΔnudJ (blue) were significantly less fit than those accessed by the wildtype (orange) & both strains accessed mutants less fit than one would expect by chance (black: all mutations occurring at =rates): spontaneous mutation is not optimised for future fitness. 11/13
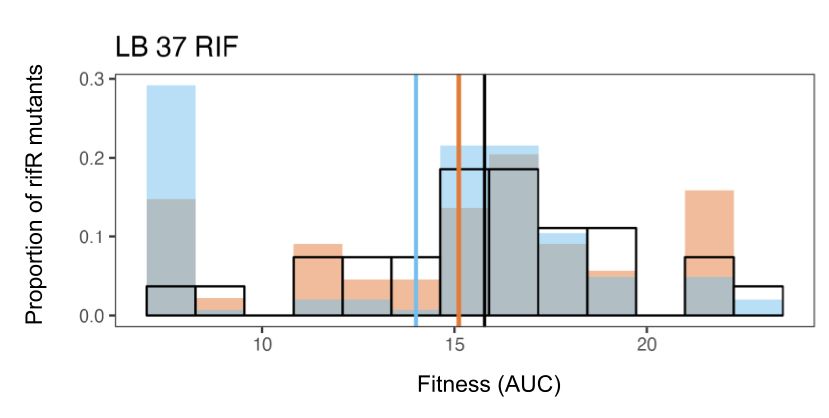
August 18, 2025 at 3:53 PM
Resistance mutations accessed by ΔnudJ (blue) were significantly less fit than those accessed by the wildtype (orange) & both strains accessed mutants less fit than one would expect by chance (black: all mutations occurring at =rates): spontaneous mutation is not optimised for future fitness. 11/13
Rather than looking at a genome-wide scale we took rifampicin resistance mutants observed in each genetic background and measured their fitness in a common genetic background. This choice to consider fitness at a single locus is reflective of resistance evolution to many antibiotics. 10/13
August 18, 2025 at 3:51 PM
Rather than looking at a genome-wide scale we took rifampicin resistance mutants observed in each genetic background and measured their fitness in a common genetic background. This choice to consider fitness at a single locus is reflective of resistance evolution to many antibiotics. 10/13
Now for the cool evolution part: work from @deepaagashe.bsky.social and others has shown that shifts in the mutational spectrum can lead to genome wide changes in the fitness effects of accessed mutations. Was our ΔnudJ strain accessing mutations with different fitness effects to the wildtype? 9/13
August 18, 2025 at 3:50 PM
Now for the cool evolution part: work from @deepaagashe.bsky.social and others has shown that shifts in the mutational spectrum can lead to genome wide changes in the fitness effects of accessed mutations. Was our ΔnudJ strain accessing mutations with different fitness effects to the wildtype? 9/13
A mutation rates is made up from the rates of many different types of mutations (the mutational spectrum). We find that ΔnudJ has a significantly altered mutational spectrum to the wildtype, e.g. in the wt 22% of mutations are A>G, in ΔnudJ this is 9%. This is reversed by IS1 mutation in waaZ. 8/13

August 18, 2025 at 3:49 PM
A mutation rates is made up from the rates of many different types of mutations (the mutational spectrum). We find that ΔnudJ has a significantly altered mutational spectrum to the wildtype, e.g. in the wt 22% of mutations are A>G, in ΔnudJ this is 9%. This is reversed by IS1 mutation in waaZ. 8/13
The antimutator ΔnudJ not only reduces mutation rates but also decreases the responsiveness of mutation rates to population density (usually mutation rates are elevated at low population densities in batch culture). Both of these traits are reverted by our strains carrying secondary mutations. 7/13

August 18, 2025 at 3:48 PM
The antimutator ΔnudJ not only reduces mutation rates but also decreases the responsiveness of mutation rates to population density (usually mutation rates are elevated at low population densities in batch culture). Both of these traits are reverted by our strains carrying secondary mutations. 7/13
I then focussed in on ΔnudJ, finding surprisingly that the mutation rate had reverted to the of the wildtype! We found that independent mutations in 2 of our stocks were responsible for this phenotype reversion, with initially frustrating inconsistencies resulting in new genetic understanding. 6/13
August 18, 2025 at 3:46 PM
I then focussed in on ΔnudJ, finding surprisingly that the mutation rate had reverted to the of the wildtype! We found that independent mutations in 2 of our stocks were responsible for this phenotype reversion, with initially frustrating inconsistencies resulting in new genetic understanding. 6/13
Disruption of nucleotide metabolism gene mutT increases mutation rates however, its sibling genes in the nudix hydrolase family had not been characterised. A massive survey by coauthor Huw Richards found that 4 knockouts from this family result in significant mutation rate decreases (nudBFJ&H) 5/13

August 18, 2025 at 3:45 PM
Disruption of nucleotide metabolism gene mutT increases mutation rates however, its sibling genes in the nudix hydrolase family had not been characterised. A massive survey by coauthor Huw Richards found that 4 knockouts from this family result in significant mutation rate decreases (nudBFJ&H) 5/13
Mutator strains in which disruption of a single gene (usually DNA replication and repair enzymes) increases mutation rates are very common in laboratory and clinical E coli populations. BUT gene disruptions that reduce mutation rates (antimutator strains) are far less frequently observed (4/13)
August 18, 2025 at 3:44 PM
Mutator strains in which disruption of a single gene (usually DNA replication and repair enzymes) increases mutation rates are very common in laboratory and clinical E coli populations. BUT gene disruptions that reduce mutation rates (antimutator strains) are far less frequently observed (4/13)
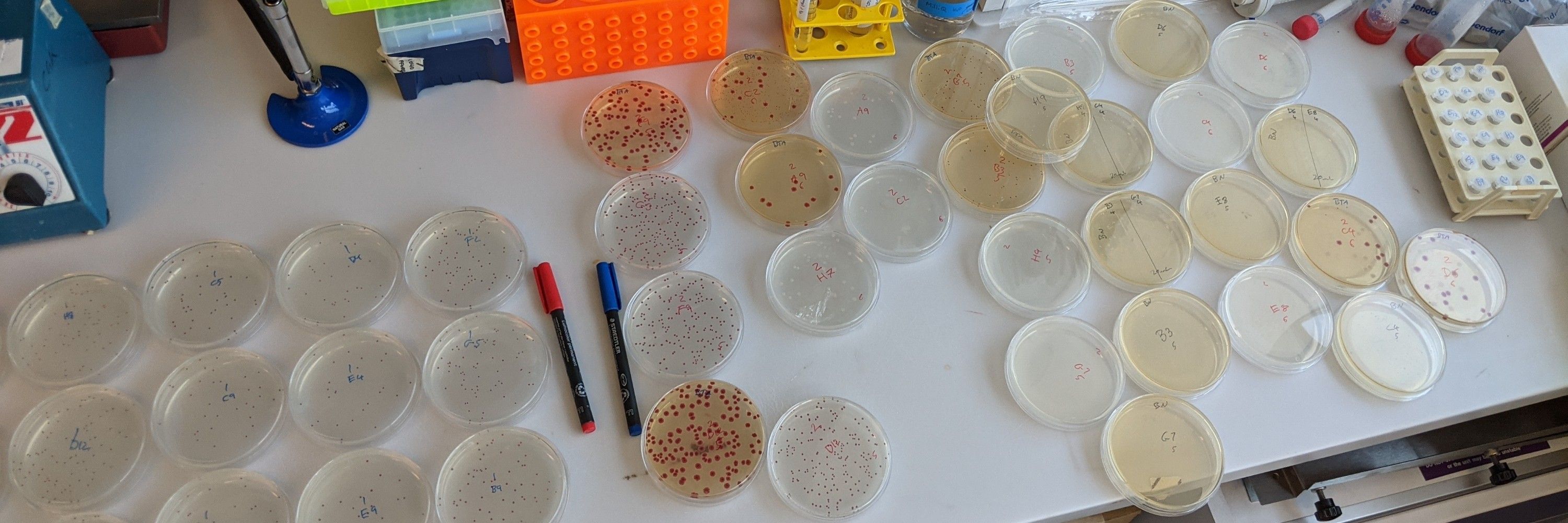
The antibiotic killed cells could stretch from Manchester to Paris and back to the UK while the total cell count would reach from my PhD lab in Manchester @mermanchester.bsky.social to my postdoc lab in Vancouver @zoology.ubc.ca 4.7 times. (3/13)
August 18, 2025 at 3:43 PM
The antibiotic killed cells could stretch from Manchester to Paris and back to the UK while the total cell count would reach from my PhD lab in Manchester @mermanchester.bsky.social to my postdoc lab in Vancouver @zoology.ubc.ca 4.7 times. (3/13)
This team effort involved 4 PhD students, 1 undergrad, 1 masters student, 6 groups leaders, 7 years of (on-and-off) research, >8.5x10^11 E. coli cells killed with antibiotics and >3.5x10^13 E. coli cells grown in total. (2/13)
August 18, 2025 at 3:41 PM
This team effort involved 4 PhD students, 1 undergrad, 1 masters student, 6 groups leaders, 7 years of (on-and-off) research, >8.5x10^11 E. coli cells killed with antibiotics and >3.5x10^13 E. coli cells grown in total. (2/13)

