Past: founder Plume Labs (acquired by AccuWeather).
Don’t be fooled by the tweets that I’ve got
I’m still I’m still @rlacombe on X (and here).
-> https://romainlacombe.com
📝 Link to paper: biorxiv.org/content/10.1...
💻 Link to code: github.com/rlacombe/Sac...

📝 Link to paper: biorxiv.org/content/10.1...
💻 Link to code: github.com/rlacombe/Sac...
@stanfordbiosci.bsky.social workshop on Experimental Design in AI for Science next week!
More info on the event here: edai4science.github.io
@stanfordbiosci.bsky.social workshop on Experimental Design in AI for Science next week!
More info on the event here: edai4science.github.io
We will likely need physics-informed models to reach accurate 3D structure predictions on rare PTMs and out-of-domain proteins beyond evolutionary priors.
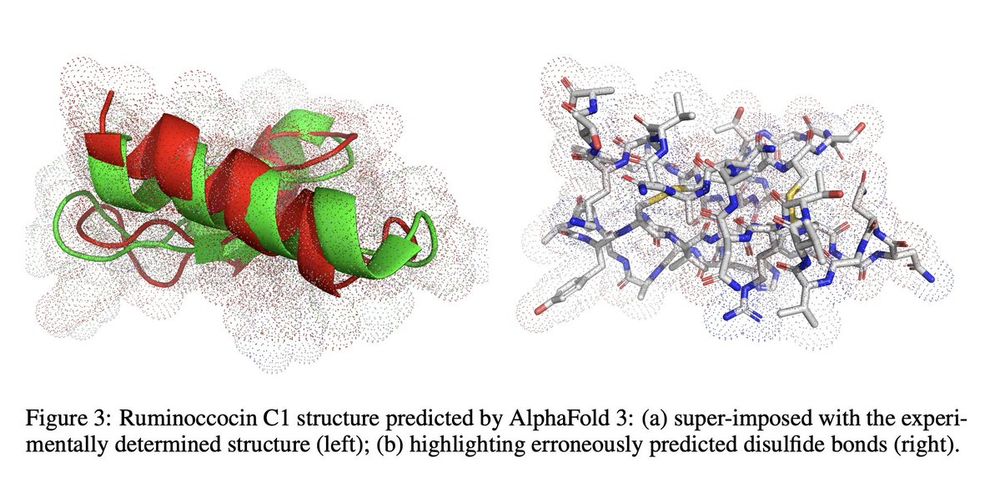
We will likely need physics-informed models to reach accurate 3D structure predictions on rare PTMs and out-of-domain proteins beyond evolutionary priors.
Most models incorrectly predicted disulfide bonds instead of thioether crosslinks, highlighting their reliance on evolutionary priors.
Most models incorrectly predicted disulfide bonds instead of thioether crosslinks, highlighting their reliance on evolutionary priors.
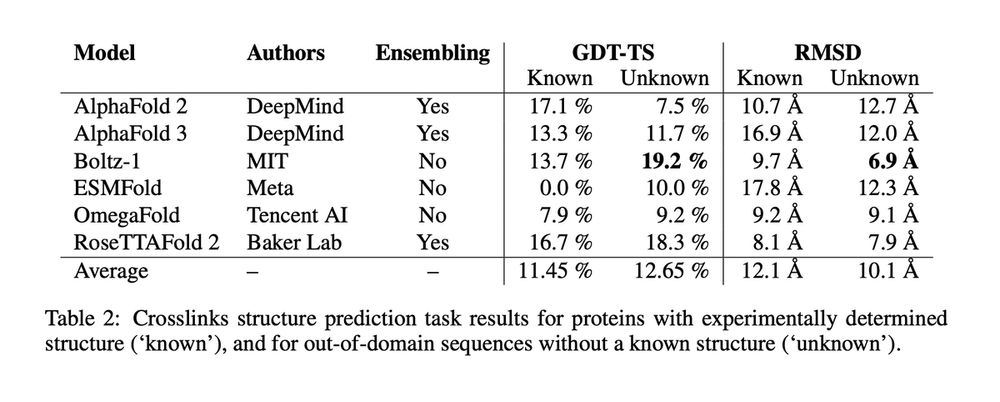
All models exhibit limited performance, with an average GDT-TS of only 11.5% for known sactipeptides and 12.6% for unknown ones.
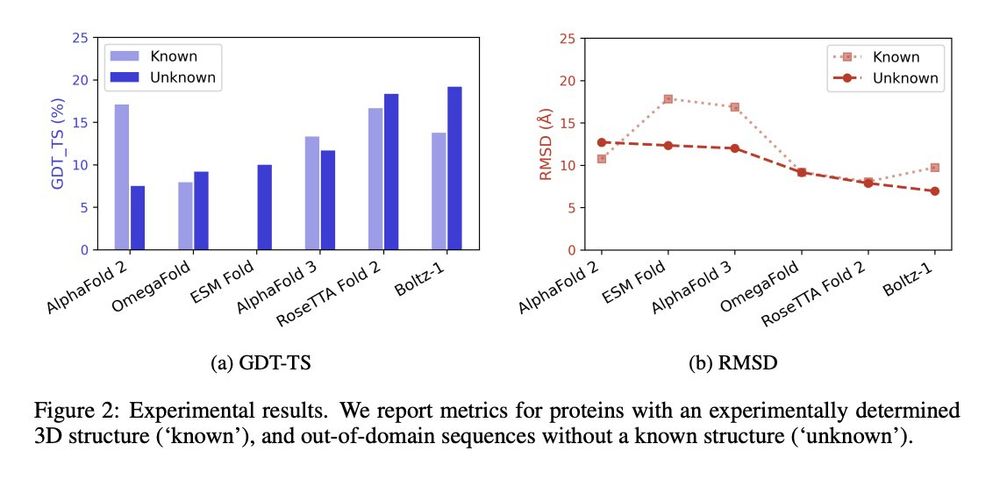
All models exhibit limited performance, with an average GDT-TS of only 11.5% for known sactipeptides and 12.6% for unknown ones.
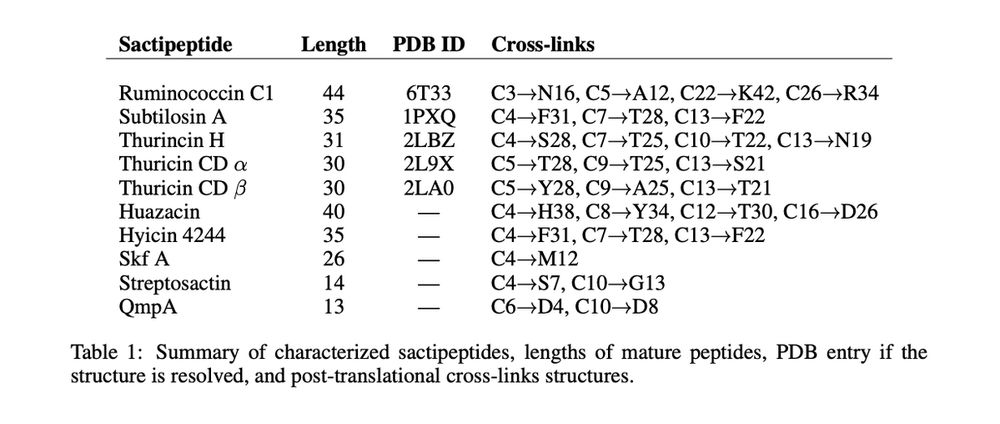
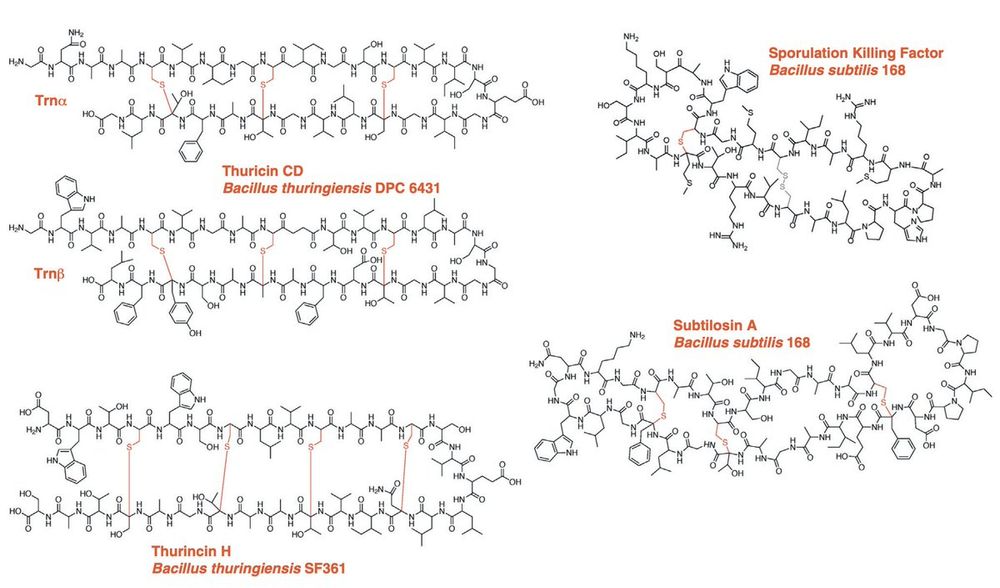
We introduce a new, zero-shot benchmark for structure models: measuring how the 3D conformations predicted for sactipeptides match the geometry imposed by their known thioether crosslinks.
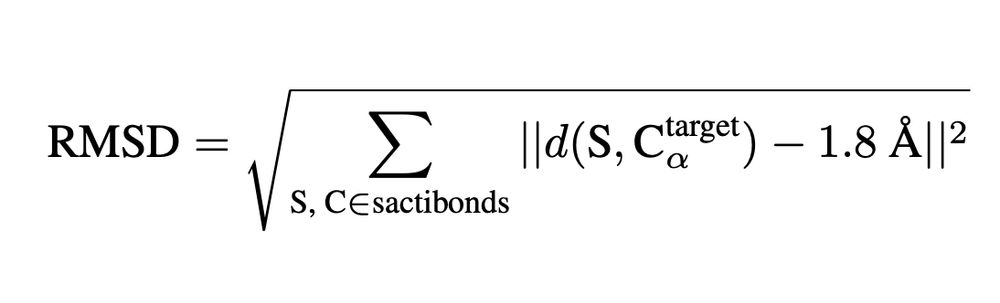
We introduce a new, zero-shot benchmark for structure models: measuring how the 3D conformations predicted for sactipeptides match the geometry imposed by their known thioether crosslinks.