
Romain Koszul
@rkoszul.bsky.social
#3Dgenome, #3R, #chromatin, #synbio, #microbiome... Opinions are my own.
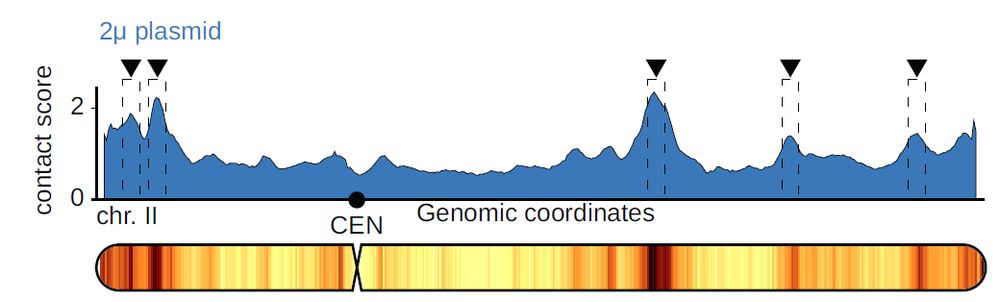
What is nice is that this phenomenon is not limited to 2u and yeast species !
Another known parasitic plasmid, Ddp5 in the amoeba Dictyostelium discoideum, also sticks to the silent regions of this genome! This suggests a more generalized phenomena.
Another known parasitic plasmid, Ddp5 in the amoeba Dictyostelium discoideum, also sticks to the silent regions of this genome! This suggests a more generalized phenomena.

March 3, 2025 at 9:24 AM
What is nice is that this phenomenon is not limited to 2u and yeast species !
Another known parasitic plasmid, Ddp5 in the amoeba Dictyostelium discoideum, also sticks to the silent regions of this genome! This suggests a more generalized phenomena.
Another known parasitic plasmid, Ddp5 in the amoeba Dictyostelium discoideum, also sticks to the silent regions of this genome! This suggests a more generalized phenomena.
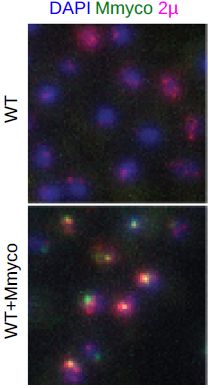
We used a yeast strain containing a non-transcribed bacterial genome to show that this artificial chromosome titrates the 2u plasmid in the “inactive” compartment
(described recently in Meneu et al www.science.org/doi/10.1126/... ) (tks Angela Taddei, Myriam Ruault & Antoine Even)
(described recently in Meneu et al www.science.org/doi/10.1126/... ) (tks Angela Taddei, Myriam Ruault & Antoine Even)
March 3, 2025 at 9:24 AM
We used a yeast strain containing a non-transcribed bacterial genome to show that this artificial chromosome titrates the 2u plasmid in the “inactive” compartment
(described recently in Meneu et al www.science.org/doi/10.1126/... ) (tks Angela Taddei, Myriam Ruault & Antoine Even)
(described recently in Meneu et al www.science.org/doi/10.1126/... ) (tks Angela Taddei, Myriam Ruault & Antoine Even)
Using dozens of published genomic datasets, and new experiments, we show that this ~6 kb molecule is positioned preferentially in untranscribed regions of the genome, often corresponding to long inactive genes.
March 3, 2025 at 9:24 AM
Using dozens of published genomic datasets, and new experiments, we show that this ~6 kb molecule is positioned preferentially in untranscribed regions of the genome, often corresponding to long inactive genes.
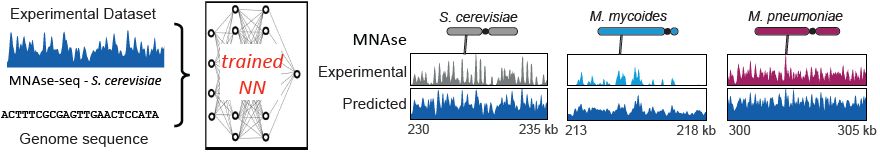
Deep learning models (@chromozz.bsky.social) trained only on yeast chromosomes predict nucleosome positioning, RNA Poll II and cohesin tracks along foreign DNA, based on the sequence alone. This implies that the behavior of any DNA in a host cell follows deterministic sequence-based rules.
February 7, 2025 at 10:22 AM
Deep learning models (@chromozz.bsky.social) trained only on yeast chromosomes predict nucleosome positioning, RNA Poll II and cohesin tracks along foreign DNA, based on the sequence alone. This implies that the behavior of any DNA in a host cell follows deterministic sequence-based rules.
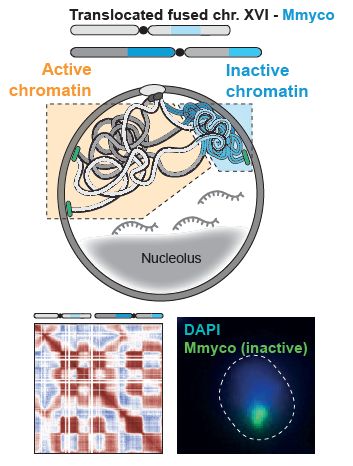
Dispersed over several chromosomes, this inactive compartment is maintained until mitosis when cohesins compact all chromosomes similarly.
Reducing transcription eliminates compartmentalization, with all yeast chromosomes now resembling the bacterial chromosome...
Reducing transcription eliminates compartmentalization, with all yeast chromosomes now resembling the bacterial chromosome...
February 7, 2025 at 10:22 AM
Dispersed over several chromosomes, this inactive compartment is maintained until mitosis when cohesins compact all chromosomes similarly.
Reducing transcription eliminates compartmentalization, with all yeast chromosomes now resembling the bacterial chromosome...
Reducing transcription eliminates compartmentalization, with all yeast chromosomes now resembling the bacterial chromosome...
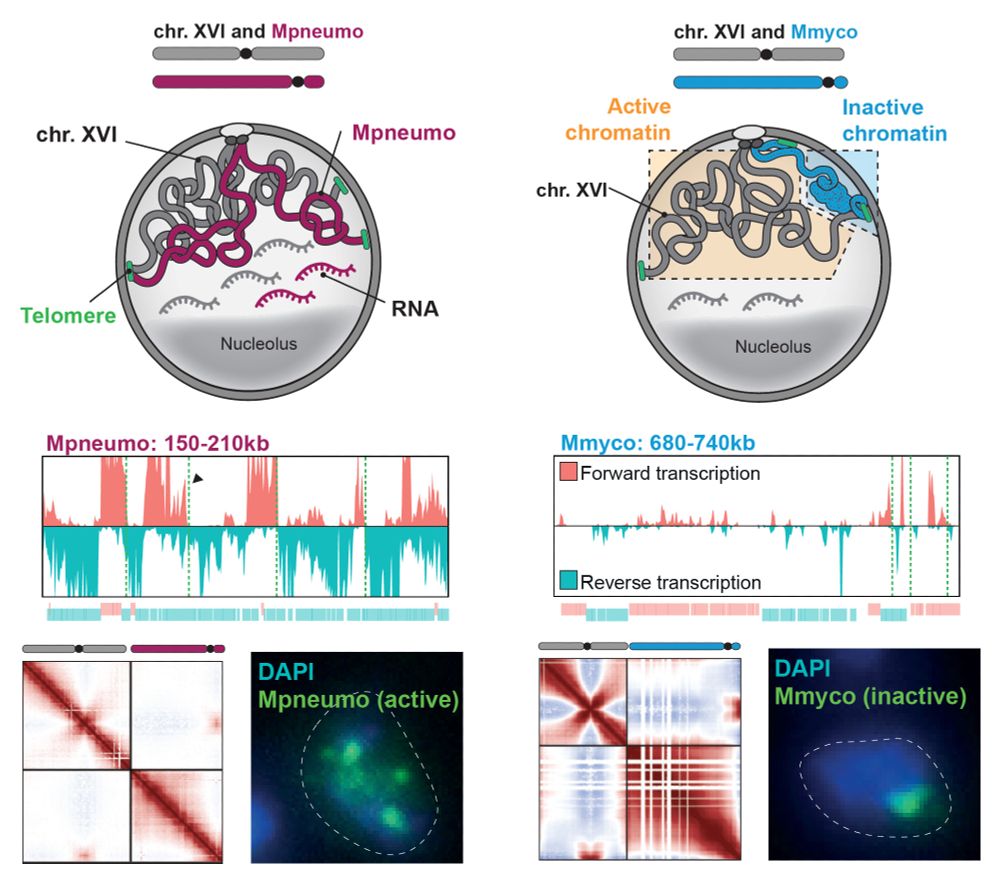
Sequences close to the yeast GC are highly transcribed, somehow following the orientation of prokaryotic genes, and mix with yeast chromosomes.
In contrast, a rich AT bacterial genome is not transcribed and forms a globule at the nuclear periphery, independently of SIR heterochromatin formation...
In contrast, a rich AT bacterial genome is not transcribed and forms a globule at the nuclear periphery, independently of SIR heterochromatin formation...
February 7, 2025 at 10:22 AM
Sequences close to the yeast GC are highly transcribed, somehow following the orientation of prokaryotic genes, and mix with yeast chromosomes.
In contrast, a rich AT bacterial genome is not transcribed and forms a globule at the nuclear periphery, independently of SIR heterochromatin formation...
In contrast, a rich AT bacterial genome is not transcribed and forms a globule at the nuclear periphery, independently of SIR heterochromatin formation...
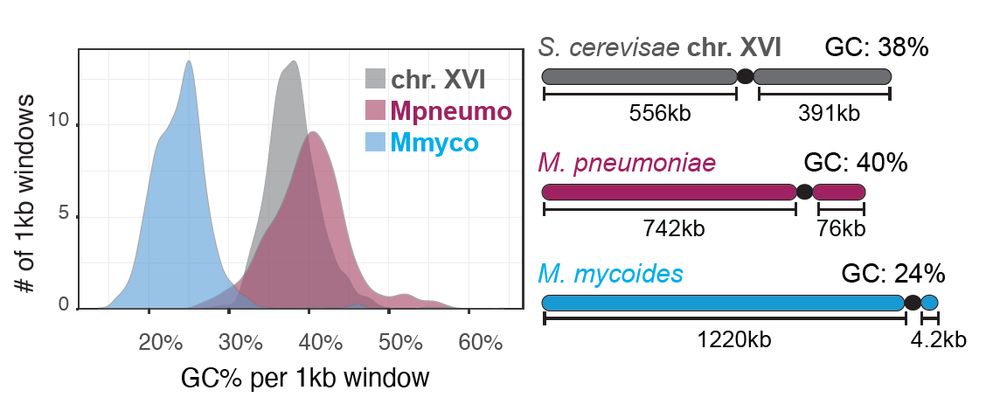
Using MNAse, ChIP, ATAC, CRAC and RNA-seq, Hi-C and imaging, we characterized the composition, activity and folding of bacterial genomes of varying sequence composition introduced as supernumerary chromosomes into budding yeast. Depending on their GC%, their fate was surprisingly different.
February 7, 2025 at 10:22 AM
Using MNAse, ChIP, ATAC, CRAC and RNA-seq, Hi-C and imaging, we characterized the composition, activity and folding of bacterial genomes of varying sequence composition introduced as supernumerary chromosomes into budding yeast. Depending on their GC%, their fate was surprisingly different.

