Energetic profiling of >110 cryo-EM amyloid fibrils (IAPP, tau, α-syn) reveals how APRs + cofactors shape fibril maturation and disease-relevant polymorphs.
Read the full paper: doi.org/10.1038/s414...

Energetic profiling of >110 cryo-EM amyloid fibrils (IAPP, tau, α-syn) reveals how APRs + cofactors shape fibril maturation and disease-relevant polymorphs.
Read the full paper: doi.org/10.1038/s414...
www.nature.com/articles/s41...
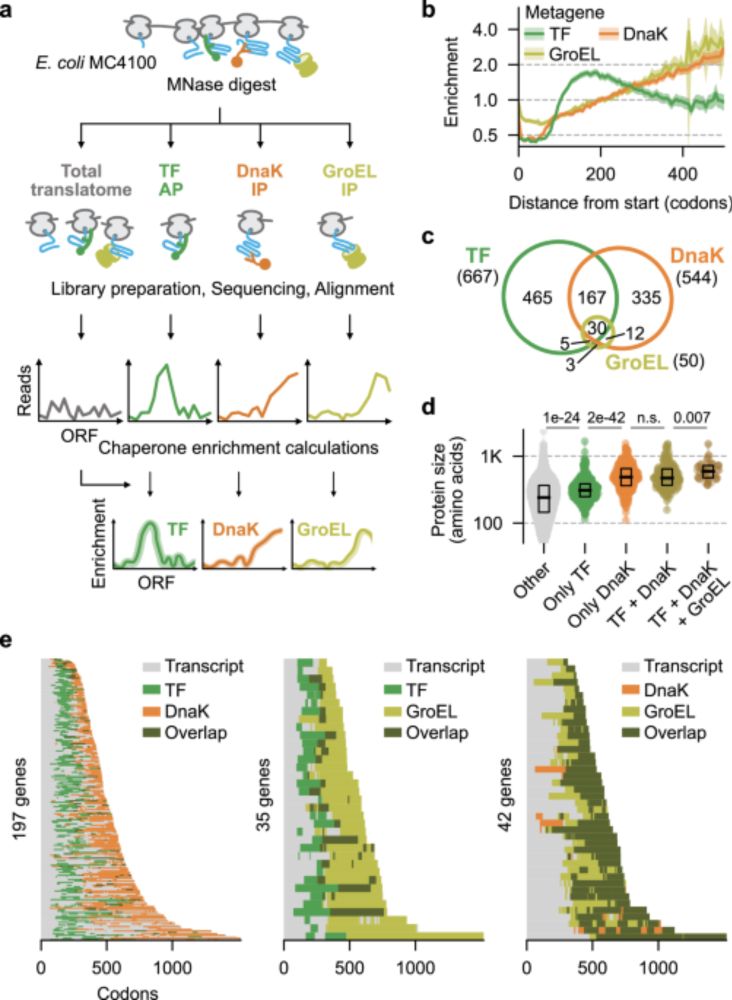
They show that bacterial chaperones bind regions of nascent proteins with exposed "unsatisfied residues" (segments missing key native contacts due to ongoing translation).
A shared theme with our recent work on Native Fold Delay.
www.nature.com/articles/s41...
They show that bacterial chaperones bind regions of nascent proteins with exposed "unsatisfied residues" (segments missing key native contacts due to ongoing translation).
A shared theme with our recent work on Native Fold Delay.
Thermodynamic profiling of cryoEM fibrils shows that key stabilizing motifs shuffle during amyloid maturation, while cofactors ease structural frustration to let energetically optimal polymorphs emerge.
www.biorxiv.org/content/10.1...

Thermodynamic profiling of cryoEM fibrils shows that key stabilizing motifs shuffle during amyloid maturation, while cofactors ease structural frustration to let energetically optimal polymorphs emerge.
www.biorxiv.org/content/10.1...
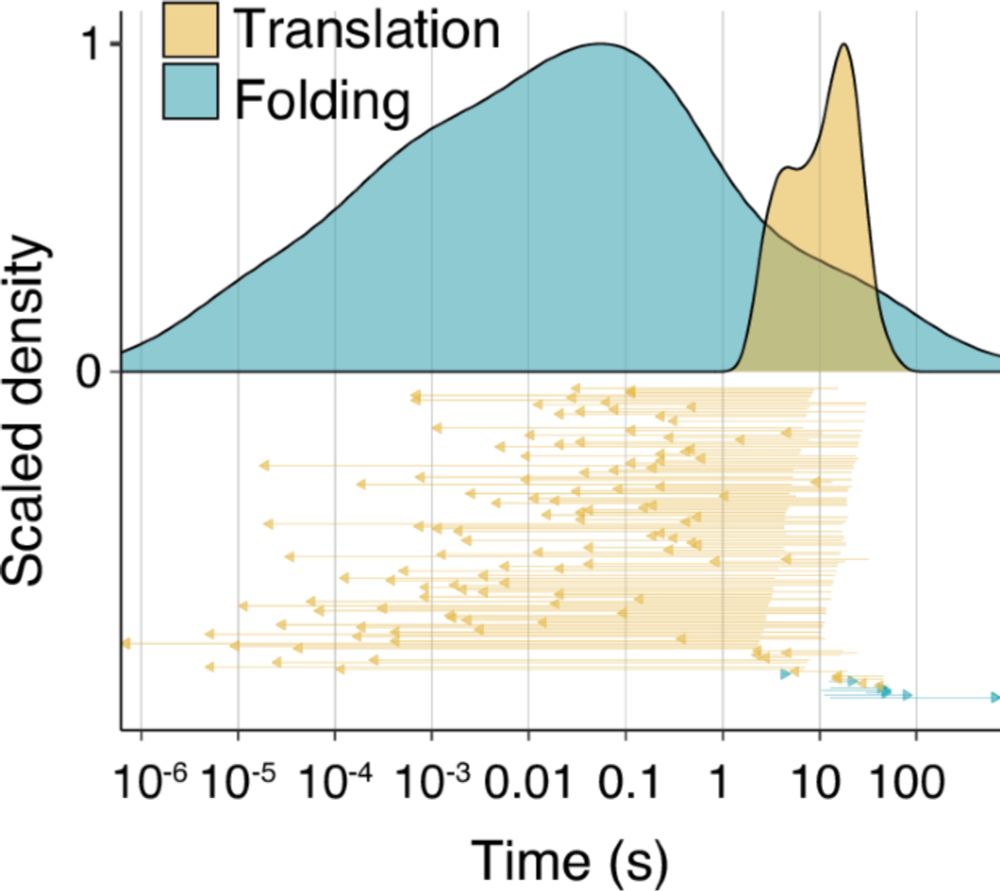
Could this be used as tool to induce translation pausing? 👀
#translation

Could this be used as tool to induce translation pausing? 👀
#translation
#ProteinTranslation #EMBORiboProf

#ProteinTranslation #EMBORiboProf
Read more here www.nature.com/articles/s41...
Read more here www.nature.com/articles/s41...

