
He/him/his

Here’s a cute cat picture for your trouble

Here’s a cute cat picture for your trouble




Register: https://tinyurl.com/MSKBridgeInfoSession2023
#PrePhD #PreMDPhD #AcademicTwitter

Register: https://tinyurl.com/MSKBridgeInfoSession2023
#PrePhD #PreMDPhD #AcademicTwitter
- easy forecasting of NPI change impacts
- data-driven
- often sig. better, never sig. worse at death forecasts than all others, incl expert-driven ones
P: https://doi.org/10.1093/jamia/ocac160
G:...

- easy forecasting of NPI change impacts
- data-driven
- often sig. better, never sig. worse at death forecasts than all others, incl expert-driven ones
P: https://doi.org/10.1093/jamia/ocac160
G:...
PRIESSTESS 🧙♀️
- fits RNA sequence and structure models
- is trivially interpretable
- generalizes as well or better as black-box...

PRIESSTESS 🧙♀️
- fits RNA sequence and structure models
- is trivially interpretable
- generalizes as well or better as black-box...

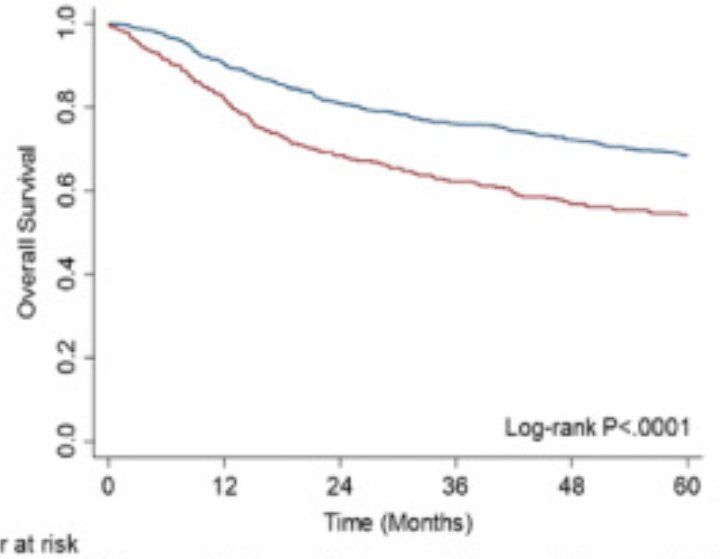
But no single feature explains the recurrence.

But no single feature explains the recurrence.




Check it out: https://www.genemania.org

Check it out: https://www.genemania.org










