
http://koo-lab.github.io


, a former postdoc who was jointly advised by me and Justin Kinney at CSHL, and also in collaboration with David McCandlish @TheDMMcC
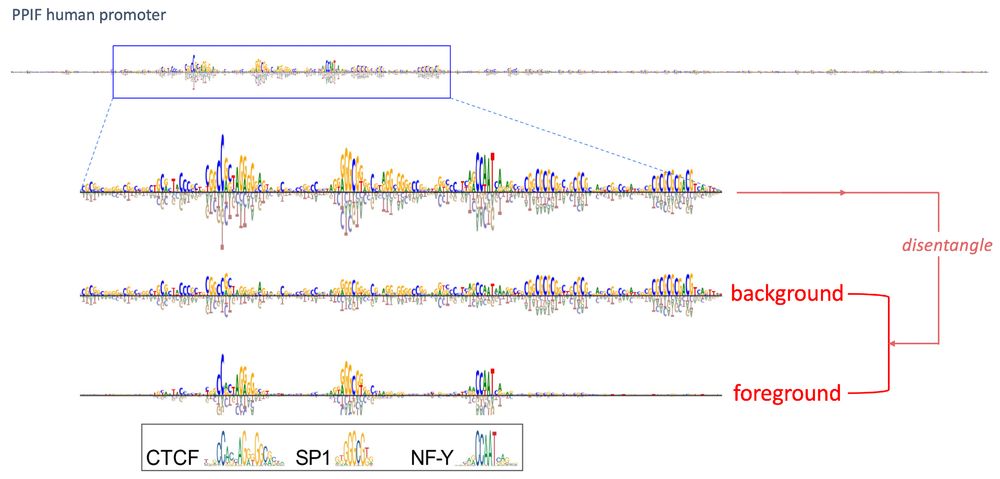
It's a beautiful followup to SQUID, our surrogate modeling approach to interpret genomic DNNs!
www.nature.com/articles/s42...

, a former postdoc who was jointly advised by me and Justin Kinney at CSHL, and also in collaboration with David McCandlish @TheDMMcC
It's a beautiful followup to SQUID, our surrogate modeling approach to interpret genomic DNNs!
www.nature.com/articles/s42...
Code: github.com/evanseitz/se...
Paper: www.biorxiv.org/content/10.1...

Code: github.com/evanseitz/se...
Paper: www.biorxiv.org/content/10.1...

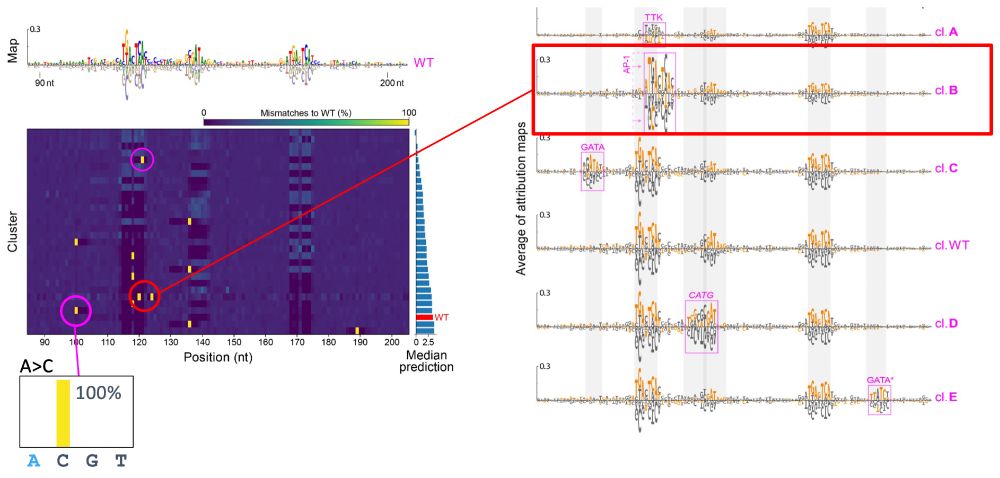
- single mutations -> no change
- double mutation -> CAAT box + new Inr
8/N

- single mutations -> no change
- double mutation -> CAAT box + new Inr
8/N




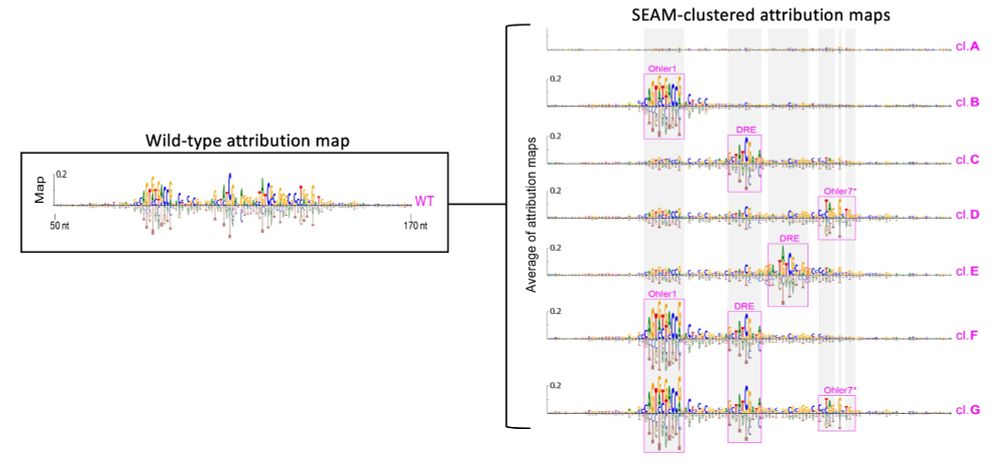
1) sample in a local region of sequence space via partial random mutagenesis
2) calculate attr maps to unveil the mechanisms
3) cluster attr maps based on shared mechanisms
4) cluster-based sequence analysis
2/N

1) sample in a local region of sequence space via partial random mutagenesis
2) calculate attr maps to unveil the mechanisms
3) cluster attr maps based on shared mechanisms
4) cluster-based sequence analysis
2/N




