
Too early to explore the Galaxy
Just in time to model the cell and climb some rocks
PhD @ UofT and Vector Institute
🔗See the call below:
joeybose.github.io/phd-positions/
✨ And a light expression of interest: forms.gle/FpgTiuatz9ft...
🔗See the call below:
joeybose.github.io/phd-positions/
✨ And a light expression of interest: forms.gle/FpgTiuatz9ft...
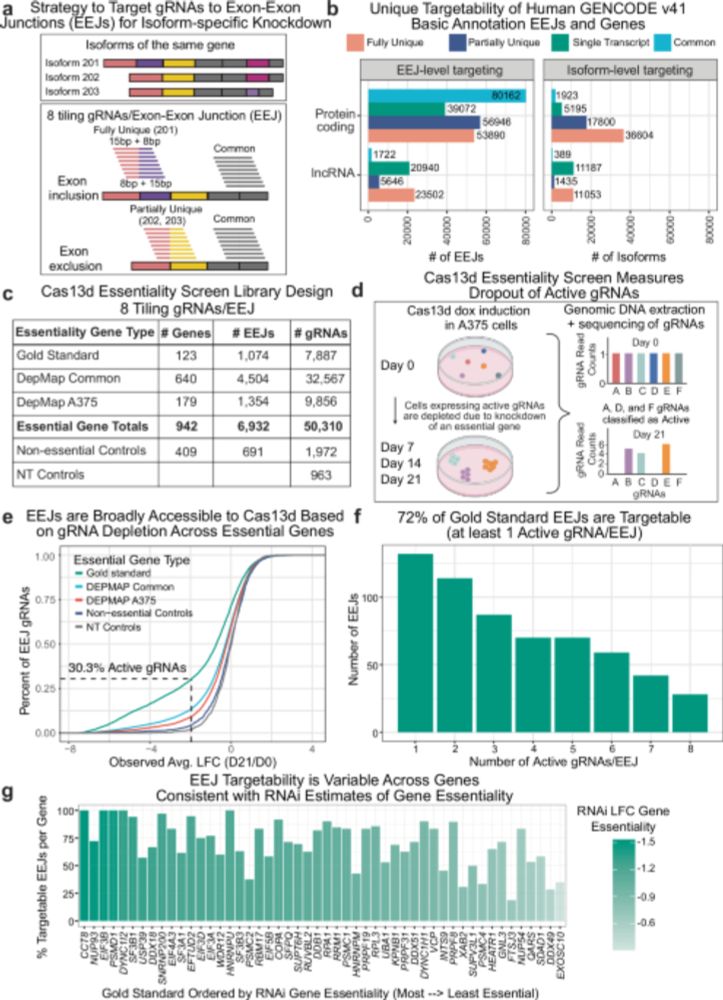
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench
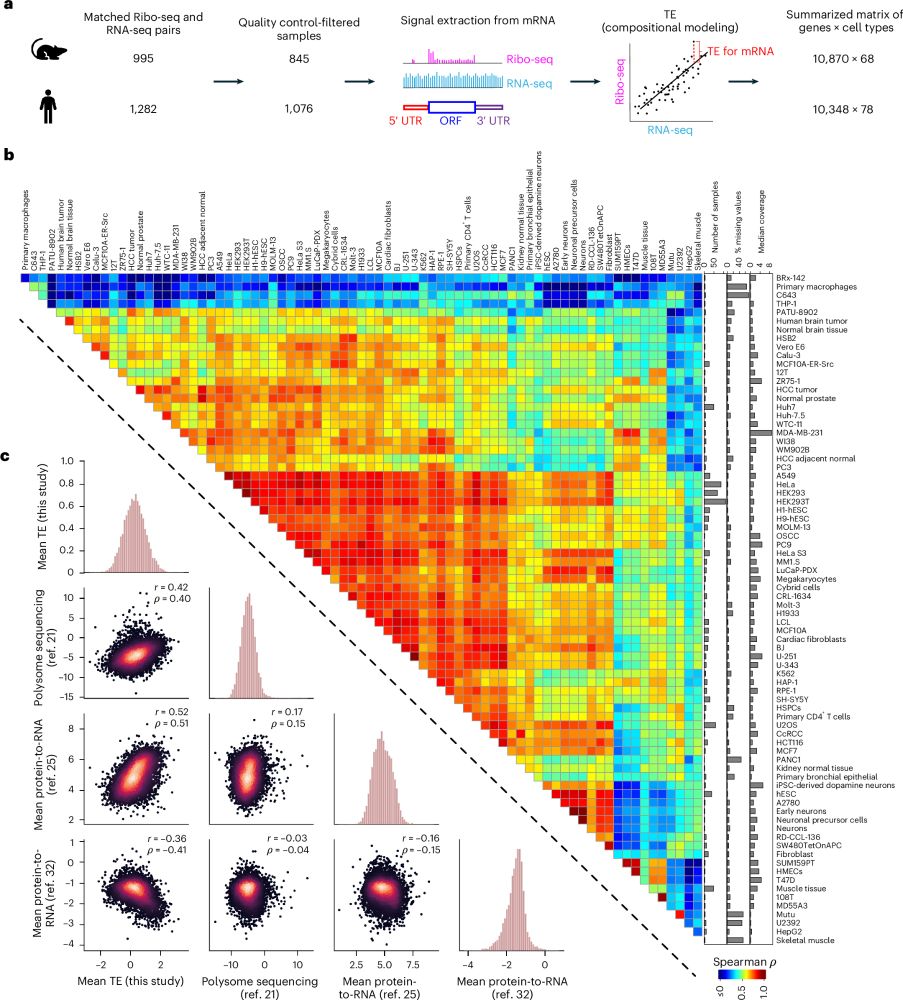
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench
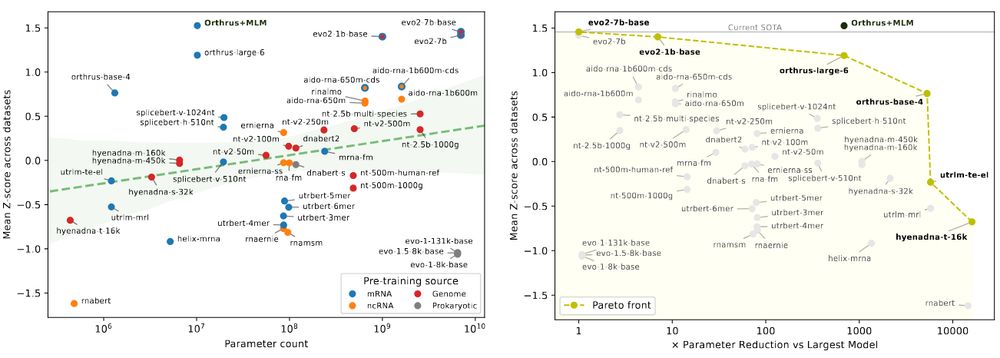
Paper: doi.org/10.1101/2025...
Code: github.com/morrislab/mR...
A 🧵

Paper: doi.org/10.1101/2025...
Code: github.com/morrislab/mR...
A 🧵

DAMUTA has independent Damage and Misrepair signatures whose activities are more interpretable and more predictive of DNA repair defects, than COSMIC SBS signatures 🧬🖥️🧪
www.biorxiv.org/content/10.1...

DAMUTA has independent Damage and Misrepair signatures whose activities are more interpretable and more predictive of DNA repair defects, than COSMIC SBS signatures 🧬🖥️🧪
www.biorxiv.org/content/10.1...
Spent the weekend reading the paper and sorting through the intuitions. Here's a visual guide and the main intuitions to understand the model and the process that created it.
newsletter.languagemodels.co/p/the-illust...
Spent the weekend reading the paper and sorting through the intuitions. Here's a visual guide and the main intuitions to understand the model and the process that created it.
newsletter.languagemodels.co/p/the-illust...
Registration open www.rna-ai.org
@hagentilgner.bsky.social @quaidmorris.bsky.social
Registration open www.rna-ai.org
@hagentilgner.bsky.social @quaidmorris.bsky.social
We had so much fun working on this with Puria (co-first author), @karush17.bsky.social, Frederik and co-supervised by Maciej and @dom-beaini.bsky.social
arxiv.org/abs/2409.08302
@valenceai.bsky.social
We had so much fun working on this with Puria (co-first author), @karush17.bsky.social, Frederik and co-supervised by Maciej and @dom-beaini.bsky.social
arxiv.org/abs/2409.08302
@valenceai.bsky.social
Come chat about a new approach to mRNA representation learning!

Come chat about a new approach to mRNA representation learning!
1. MolPhenix (main track): Multi-modal learning learning joint representations between molecular structures & phenomic data
1. MolPhenix (main track): Multi-modal learning learning joint representations between molecular structures & phenomic data

