@cp-cellreports.bsky.social @cellpress.bsky.social

@cp-cellreports.bsky.social @cellpress.bsky.social
@cp-cellreports.bsky.social
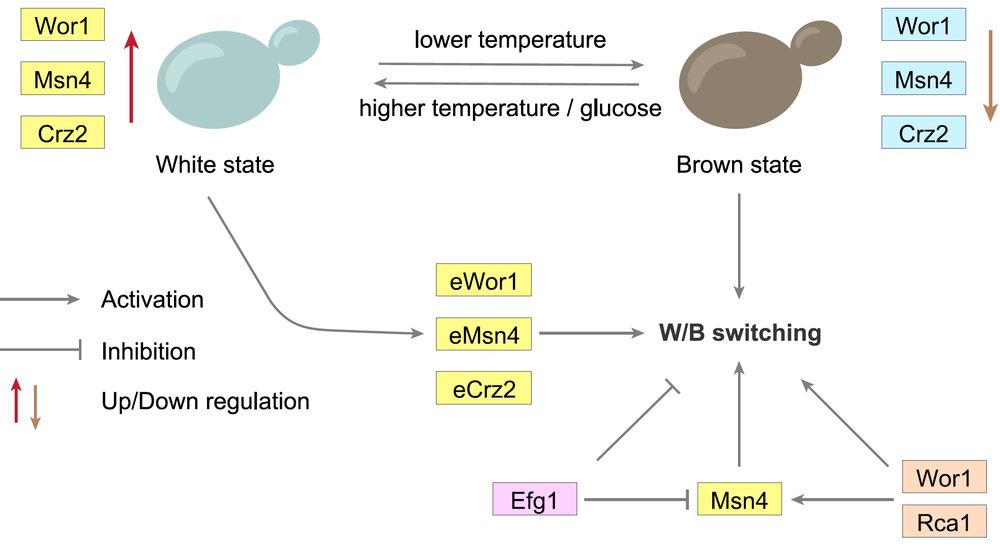
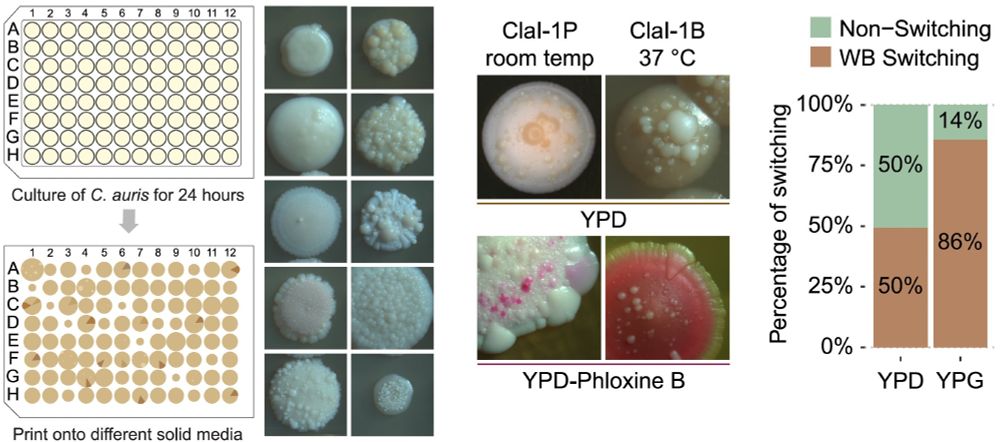
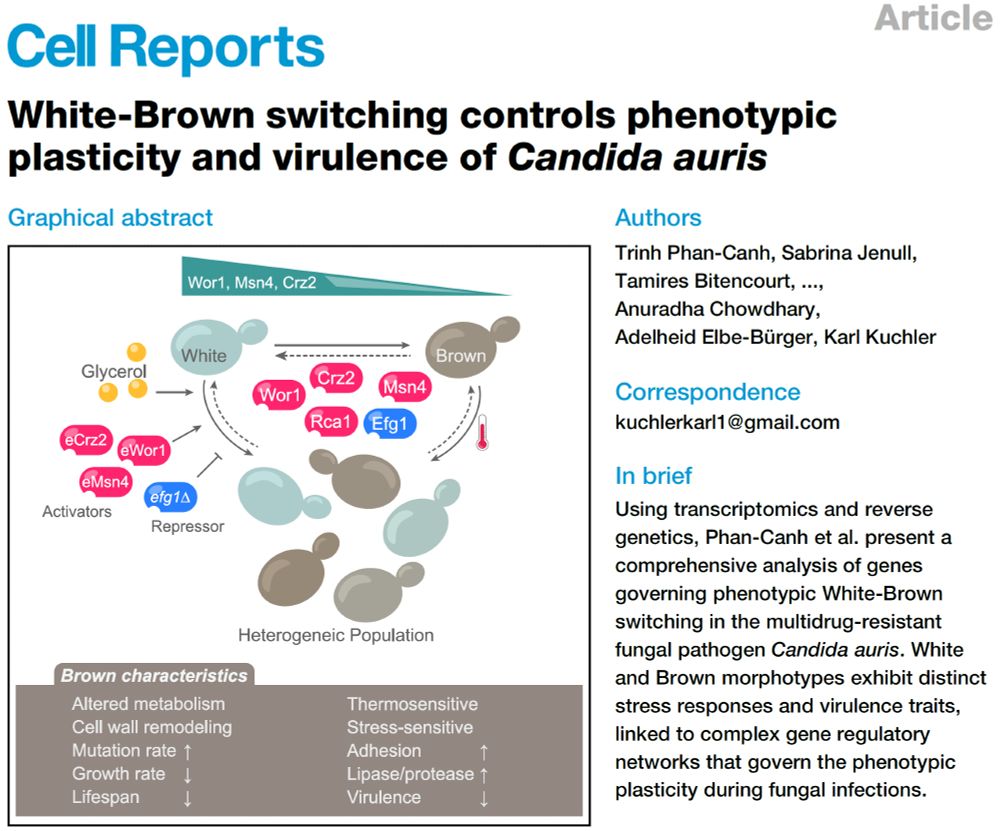
: White–Brown switching in #Candida #auris is now out! This may be the first comprehensive study of epigenetic switching in #auris, shedding light on clinical variability in virulence and antifungal resistance. Link in comment! 1/n
@cp-cellreports.bsky.social
: White–Brown switching in #Candida #auris is now out! This may be the first comprehensive study of epigenetic switching in #auris, shedding light on clinical variability in virulence and antifungal resistance. Link in comment! 1/n
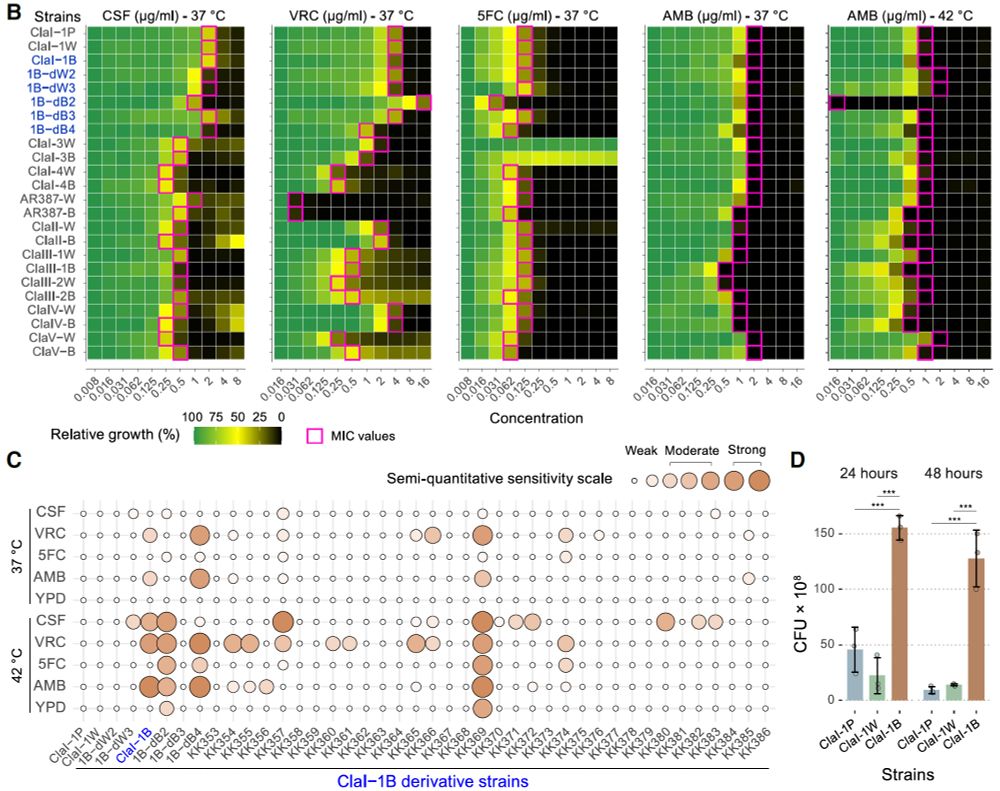
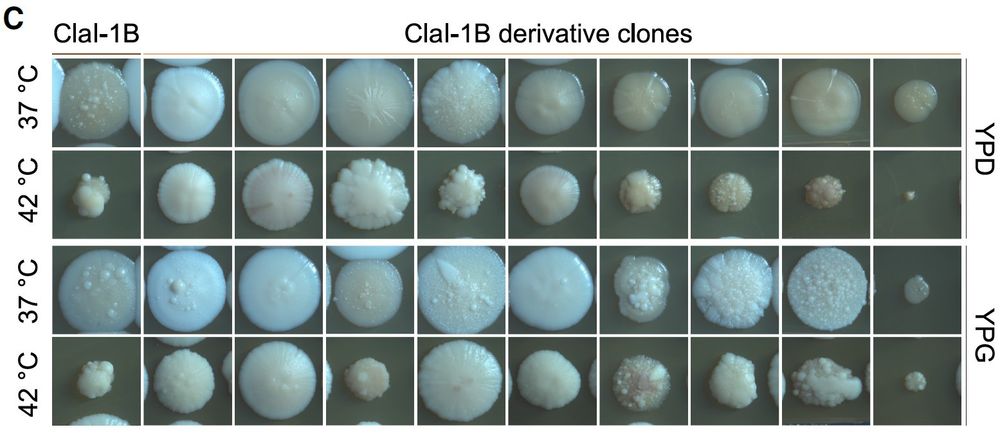
Check it out: Rapid in vitro evolution of flucytosine resistance in Candida auris
1/4
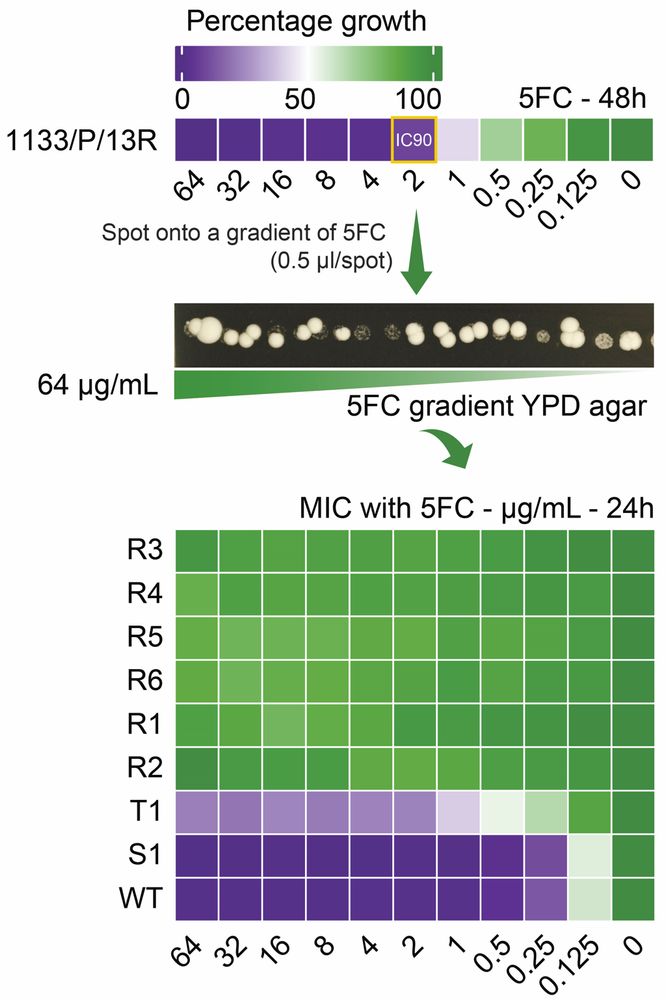
Check it out: Rapid in vitro evolution of flucytosine resistance in Candida auris
1/4
“HDAC1 Fine-Tunes Th17 Polarization In Vivo to Restrain Tissue Damage in Fungal Infections”
www.cell.com/cell-reports...
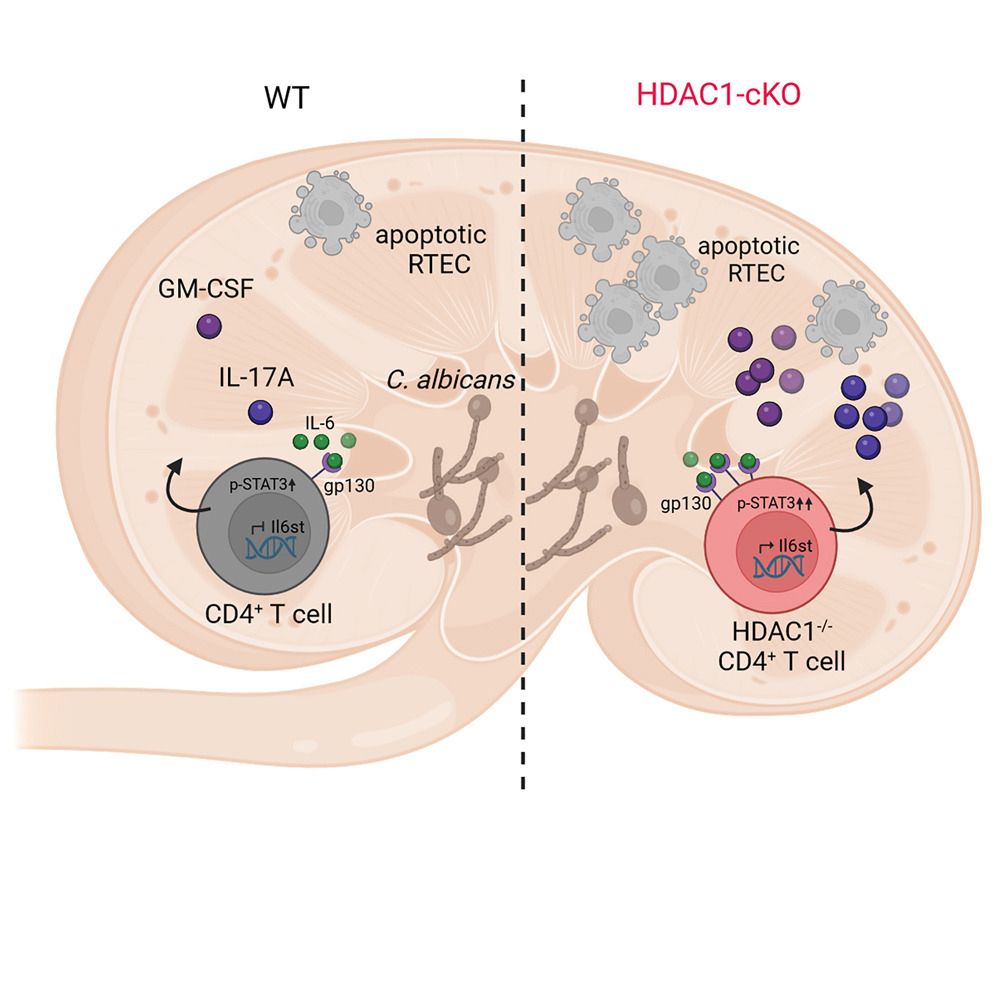
“HDAC1 Fine-Tunes Th17 Polarization In Vivo to Restrain Tissue Damage in Fungal Infections”
www.cell.com/cell-reports...
Paper: www.nature.com/articles/s42...
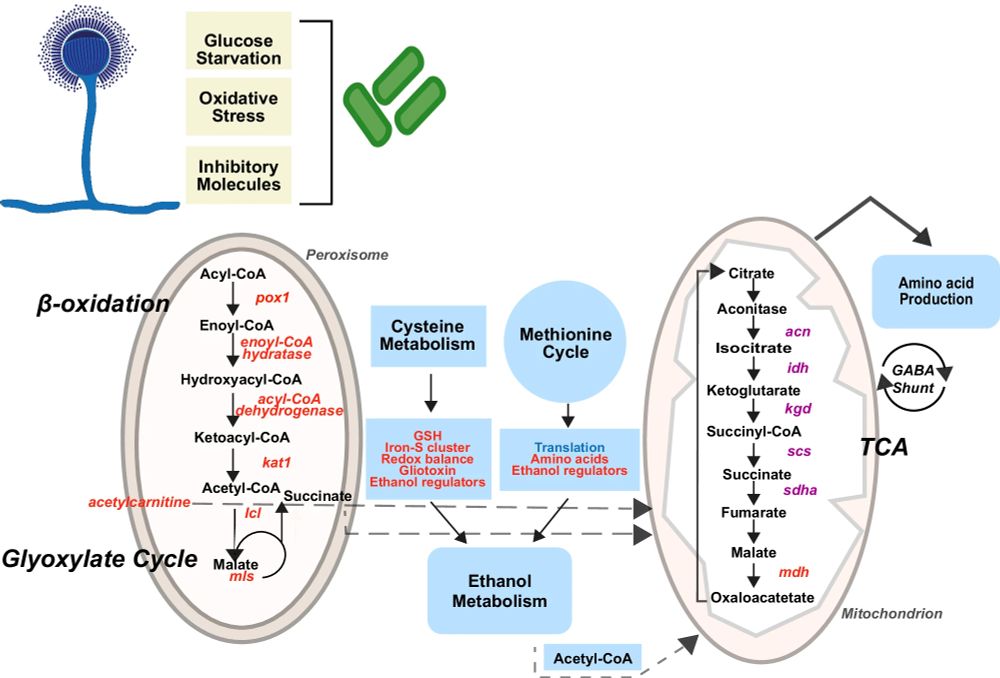
Paper: www.nature.com/articles/s42...

PLOS Pathogens: journals.plos.org/plospathogen...
#Candidaauris #Candida #infectiousdisease #morphogenesis #climatechange

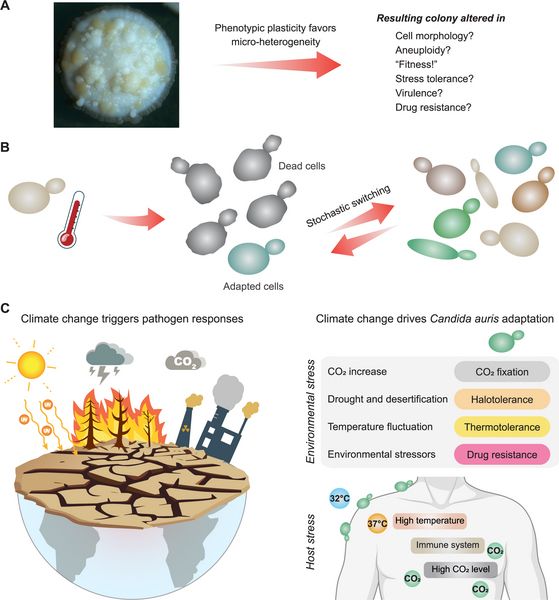
PLOS Pathogens: journals.plos.org/plospathogen...
#Candidaauris #Candida #infectiousdisease #morphogenesis #climatechange

